Plasmodium falciparum biology: Difference between revisions
| Line 1: | Line 1: | ||
{{DISPLAYTITLE:''Plasmodium falciparum'' biology}} |
|||
{{Taxobox |
{{Taxobox |
||
|name=''Plasmodium falciparum'' |
|||
| color = khaki |
|||
|image=Plasmodium_falciparum_01.png |
|||
| name = ''Plasmodium falciparum'' |
|||
|image_width=240px |
|||
| image = Plasmodium_falciparum_01.png |
|||
|image_caption=Blood smear with ''Plasmodium falciparum'' |
|||
| image_width = 240px |
|||
|domain=[[Eukaryota]] |
|||
| image_caption = Blood smear of ''Plasmodium falciparum'' |
|||
| |
|regnum=[[Chromalveolata]] |
||
| |
|superphylum=[[Alveolata]] |
||
| |
|phylum=[[Apicomplexa]] |
||
| |
|classis=[[Aconoidasida]] |
||
| |
|ordo=[[Haemosporida]] |
||
|familia=[[Plasmodiidae]] |
|||
| genus = ''[[Plasmodium]]'' |
|||
|genus=''[[Plasmodium]]'' |
|||
| species = '''''P. falciparum''''' |
|||
| |
|species='''''P. falciparum''''' |
||
|binomial=''[[Plasmodium falciparum]]'' |
|||
| binomial_authority = [[William Henry Welch|Welch]], 1897 |
|||
|binomial_authority=[[William Henry Welch|Welch]], 1897}} |
|||
}} |
|||
{{very long|date=December 2012}} |
|||
''[[Plasmodium falciparum]]'' has been the focus of much research due to it being the causative agent of malaria. This article describes some of the recent findings surrounding the unique biology of this organism. |
|||
{{split-apart|date=December 2012}} |
|||
==Life Cycle== |
|||
''[[Plasmodium falciparum]]'' has been the focus of much research due to it being the causative agent of [[malaria]]. This article describes some of the recent findings surrounding the unique biology of this organism. |
|||
==Overview of life cycle== |
|||
''[[Plasmodium falciparum]]'' has a complicated life-cycle, requiring both a human and a [[mosquito]] host, and differentiating multiple times during its transmission/infection process.<ref name="wirth">{{cite journal |
''[[Plasmodium falciparum]]'' has a complicated life-cycle, requiring both a human and a [[mosquito]] host, and differentiating multiple times during its transmission/infection process.<ref name="wirth">{{cite journal |
||
|last=Wirth |
|||
|first=Dyann |
|||
|title=The parasite genome: Biological revelations |
|||
| authorlink = |
|||
|journal=Nature |
|||
| coauthors = |
|||
|volume=419 |
|||
| title = The parasite genome: Biological revelations |
|||
|issue=6906 |
|||
| journal = Nature |
|||
|pages=495–496 |
|||
| volume = 419 |
|||
|date=3 October 2002 |
|||
| issue = |
|||
|url=http://www.nature.com/nature/journal/v419/n6906/full/419495a.html |
|||
| pages = 495–496 |
|||
|doi=10.1038/419495a |
|||
| publisher = |
|||
|pmid=12368862}}</ref> |
|||
| location = |
|||
| date = 3 October 2002 |
|||
| url = http://www.nature.com/nature/journal/v419/n6906/full/419495a.html |
|||
| doi =10.1038/419495a |
|||
| id = |
|||
| accessdate = }}</ref> |
|||
[[Image:Plasmodium lifecycle PHIL 3405 lores.jpg|frame|center|Plasmodium life cycle<ref name="CDC-DPD">{{cite web |
[[Image:Plasmodium lifecycle PHIL 3405 lores.jpg|frame|center|Plasmodium life cycle<ref name="CDC-DPD">{{cite web |
||
|title=DPDx - Malaria Image Library |
|||
| last = |
|||
|url=http://www.dpd.cdc.gov/dpdx/HTML/ImageLibrary/Malaria_il.htm}}</ref>]] |
|||
| first = |
|||
| authorlink = |
|||
==Genome== |
|||
| coauthors = |
|||
| title = DPDx - Malaria Image Library |
|||
The genome of ''[[Plasmodium falciparum]]'' (clone 3D7) was fully sequenced in 2002.<ref name="gardner"/> The parasite has a 23 megabase genome, divided into 14 [[chromosomes]].<ref name="gardner"/> The genome codes for approximately 5,300 genes. About 60% of the putative proteins have little or no similarity to proteins in other organisms and thus currently have no functional assignment.<ref name="gardner">{{cite journal |
|||
| work = |
|||
|last=Gardner |
|||
| publisher = |
|||
|first=Malcolm |
|||
| date = |
|||
|title=Genome sequence of the human malaria parasite ''Plasmodium falciparum'' |
|||
| url = http://www.dpd.cdc.gov/dpdx/HTML/ImageLibrary/Malaria_il.htm |
|||
|journal=Nature |
|||
| format = |
|||
|volume=419 |
|||
| doi = |
|||
|issue=6906 |
|||
| accessdate = }}</ref>]] |
|||
|pages=498–511 |
|||
===Human infection=== |
|||
|date=3 October 2002 |
|||
''[[Plasmodium falciparum|P. falciparum]]'' is transmitted to humans by the females of the ''[[Anopheles]]'' species of [[mosquito]]. There are about 460 species of ''[[Anopheles]]'' [[mosquito]], but only 68 transmit malaria. ''[[Anopheles gambiae]]'' is one of the best malaria vectors since it is long-lived, prefers feeding on humans, and lives in areas near human habitation. ''[[Anopheles gambiae|A. gambiae]]'' is found in Africa. <ref name="1.3">{{cite web |
|||
|url=http://www.nature.com/nature/journal/v419/n6906/full/nature01097.html |
|||
| last = |
|||
|doi=10.1038/nature01097 |
|||
| first = |
|||
|pmid=12368864 |
|||
| authorlink = |
|||
|last2=Hall |
|||
| coauthors = |
|||
|first2=N |
|||
| title = Malaria eModule - Transmission |
|||
|last3=Fung |
|||
| work = |
|||
|first3=E |
|||
| publisher = |
|||
|last4=White |
|||
| date = |
|||
|first4=O |
|||
| url = http://www.impact-malaria.com/FR/EPS/Formations_et_cours_internationaux/Formation_de_la_Liverpool_School_LSTMH/cours_liverpool/Unit_1/1_3.html |
|||
|last5=Berriman |
|||
| format = |
|||
|first5=M |
|||
| doi = |
|||
|last6=Hyman |
|||
| accessdate = }}</ref> |
|||
|first6=RW |
|||
|last7=Carlton |
|||
|first7=JM |
|||
|last8=Pain |
|||
|first8=A |
|||
|last9=Nelson |
|||
|first9=KE |
|||
|first10=Sharen |
|||
|first11=Ian T. |
|||
|first12=Keith |
|||
|first13=Jonathan A. |
|||
|first14=Kim |
|||
|first15=Steven L. |
|||
|first16=Alister |
|||
|first17=Sue |
|||
|first18=Man-Suen |
|||
|first19=Vishvanath |
|||
|first20=Shamira J. |
|||
|first21=Bernard |
|||
|first22=Jeremy |
|||
|first23=Sam |
|||
|first24=Mihaela |
|||
|first25=Jonathan |
|||
|first26=Jeremy |
|||
|first27=Daniel |
|||
|first28=Michael W. |
|||
|first29=Akhil B. |
|||
|first30=David M. A. |
|||
|displayauthors=30}}</ref> It is estimated 52.6% of the genome is a coding region, with 53.9% of the putative genes containing at least one intron.<ref name="gardner"/> |
|||
===Haploid/diploid=== |
|||
It is [[haploid]] during nearly all stages of its life-cycle, except for a brief period after fertilization when it is diploid from the [[ookinete]] to sporogenic stages within the [[mosquito]] gut. |
|||
===AT richness=== |
|||
The ''[[Plasmodium falciparum|P. falciparum]]'' genome has an AT content of roughly 80.6%.<ref name="gardner"/> Within the intron and intergenic regions, this AT composition rises to roughly 90%. The putative exons contain an AT content of 76.3%. The parasite's AT content is very high in comparison to other organisms. For example, the genomes of ''[[Saccharomyces cerevisiae]]'' and ''[[Arabidopsis thaliana]]'' are considered AT rich but have AT contents of 62% and 65%, respectively.<ref name="gardner"/> |
|||
===Promoters=== |
|||
Although promoters are present in the genome, very little is known about them. |
|||
===Recombination=== |
|||
The overall recombination rate is 9.6 kilobase per [[centimorgan]] and 54 candidate recombination hotspots have been identified.<ref name="Jiang2011">Jiang H, Li N, Gopalan V, Zilversmit MM, Varma S, Nagarajan V, Li J, Mu J, Hayton K, Henschen B, Yi M, Stephens R, McVean G, Awadalla P, Wellems TE, Su XZ (2011) High recombination rates and hotspots in a Plasmodium falciparum genetic cross" ''Genome Biol'' 12(4) R33</ref> The [[centromere]]s are found in chromosome regions largely devoid of recombination activity like other organisms. Within the hotspots a number of motifs were enriched including a 12 base pair G/C-rich motif with 3 base pair periodicity that may interact with a protein containing 11 predicted [[zinc finger]] arrays. |
|||
===Subtelomeric regions=== |
|||
Throughout the eukaryotic kingdom, the overall structure of chromosome ends is conserved and is characterized by the telomeric tract - a series of short G-rich repeats. This is succeeded by an extensive subtelomeric region consisting of various types and lengths of repeats — the telomere associated sequences (TAS).<ref name=Pryde1997>Pryde FE, Gorham HC, Louis EJ (1997) Chromosome ends: all the same under their caps. ''Curr Opin Genet Dev'' 7(6) 822-828</ref> In general transcription of genes located next to telomeres is repressed, a phenomenon termed the telomere position effect. This effect is somewhat misnamed as it appears to be due to the sequences found in this region rather than the position of the gene.<ref name=Stavenhagen1994>{{cite journal |author=Stavenhagen JB, Zakian VA |year=1994 |title=Internal tracts of telomeric DNA act as silencers in ''Saccharomyces cerevisiae'' |journal=Genes Dev |volume=8 |issue=12 |pages=1411–1422 |doi=10.1101/gad.8.12.1411 |pmid=7926741}}</ref> |
|||
Subtelomeric regions in general are low in gene density, low in transcription, low in recombination, late replicating, are involved in protecting the end from degradation and end-to-end fusions and in completing replication. The subtelomeric repeats can rescue chromosome ends when [[telomerase]] fails, buffer subtelomerically located genes against transcriptional silencing and protect the genome from deleterious rearrangements due to ectopic recombination. They may also be involved in fillers for increasing chromosome size to some minimum threshold level necessary for chromosome stability; act as barriers against transcriptional silencing; provide a location for the adaptive amplification of genes; and be involved in secondary mechanism of telomere maintenance via recombination when telomerase activity is absent. The repressive [[histone]] 3 lysine 9 tri-methylation mark and [[heterochromatin protein 1]] are found throughout the TAS region and adjacent gene families on all chromosomes. These heterochromatic marks are important in telomere proximal gene silencing. |
|||
In parasitic species genes involved in antigenic variation are commonly located in these regions.<ref name=Barry2003>Barry JD, Ginger ML, Burton P, McCulloch R (2003) Why are parasite contingency genes often associated with telomeres? ''Int J Parasitol'' 33(1) 29-45</ref> |
|||
The chromsomes of ''P. falciparum'' conform to this basic pattern with the ends of the chromosomes consist of a stretch of telomeric GGGTT(T/C)A repeats with an average size of 1.2 kilobases (kb). This is followed by an extensive 20 to 40 kb TAS domain. These show a high degree of conservation within the genome and contain significant amounts of repeated structure.<ref name="gardner"/> Telomere repeats are followed by a mosaic of six distinct telomere associated repetitive elements (TAREs 1-6) which are always found in the same order but vary in size. |
|||
Many genes involved in antigenic variation are located in the subtelomeric regions of the chromosomes. These are divided into the ''var'', ''rif'', and ''stevor'' families. Within the genome, there are 59 ''var'', 149 ''rif'', and 28 ''stevor'' genes along with multiple pseudogenes and truncations.<ref name="gardner"/> |
|||
The chromosome ends form clusters of 4–7 telomeres that localize around the nuclear periphery.<ref name=Freitas-Junior2000>{{cite journal |author=Freitas-Junior LH |year=2000 |title=Frequent ectopic recombination of virulence factor genes in telomeric chromosome clusters of ''P. falciparum'' |journal=Nature |volume=407 |issue=6807 |pages=1018–1022 |author-separator=, |author2=Bottius E |author3=Pirrit LA |author4=Deitsch KW |author5=Scheidig C |author6=Guinet F |author7=Nehrbass U |author8=Wellems TE |author9=Scherf A |display-authors=9 |doi=10.1038/35039531 |pmid=11069183}}</ref> Within this location the telomeric areas undergo frequent recombination which seems to increases antigenic variation. |
|||
==Transcriptome== |
|||
[[Image:Pftranscriptome.jpg|thumb|Phaseogram of ''[[Plasmodium falciparum]]'' intraerythrocytic development cycle transcriptome<ref name="bozdech"/>]] |
|||
Transcription in ''P. falciparum'' appears to have significant differences from that found in most other eukaryotes examined to date with the chromatin undergoing dramatic unheavals during the cell cycle.<ref name="Ponts2010">Ponts N, Harris EY, Prudhomme J, Wick I, Eckhardt-Ludka C, Hicks GR, Hardiman G, Lonardi S, Le Roch KG. Nucleosome landscape and control of transcription in the human malaria parasite. Genome Res.</ref> |
|||
A transcriptome analysis has been conducted on the intraerythrocytic development cycle of ''[[Plasmodium falciparum|P. falciparum]]''.<ref name="bozdech">{{cite journal |
|||
|last=Bozdech |
|||
|first=Zbynek |
|||
|title=The Transcriptome of the Intraerythrocytic Developmental Cycle of Plasmodium falciparum |
|||
|journal=PLoS Biology |
|||
|volume=1 |
|||
|issue=1 |
|||
|pages=E5 |
|||
|date=August 18, 2003 |
|||
|url=http://biology.plosjournals.org/perlserv/?request=get-document&doi=10.1371%2Fjournal.pbio.0000005 |
|||
|doi=10.1371/journal.pbio.0000005 |
|||
|pmid=12929205 |
|||
|pmc=176545 |
|||
|last2=Llinás |
|||
|first2=Manuel |
|||
|last3=Pulliam |
|||
|first3=Brian Lee |
|||
|last4=Wong |
|||
|first4=Edith D. |
|||
|last5=Zhu |
|||
|first5=Jingchun |
|||
|last6=Derisi |
|||
|first6=Joseph L.}}</ref> Roughly 60% of the genome is transcriptionally active during this portion of the parasite's life cycle. Whereas many genes appear to have stable mRNA levels throughout the cycle, many of the genes are transcriptionally regulated in a continuous cascade. |
|||
The transition from early trophozoite to trophozoite to schizont correlates with the ordered induction of genes related to transcription/translation machinery, metabolic synthesis, energy metabolism, DNA replication, protein degradation, plastid functions, merozoite invasion, and motility. Closely adjacent genes along the chromosome do not exhibit common transcription characteristics. Thus, genes are likely individually regulated along the parasite chromosome. Conversely, the apicoplast genome is polycistronic and most of its genes are coexpressed during the intraerythrocytic development cycle.<ref name="bozdech"/> |
|||
===Introns=== |
|||
The intron splicing has been examined experimentally.<ref name=Zhang2011>{{cite journal |author=Zhang X, Tolzmann CA, Melcher M, Haas BJ, Gardner MJ, Smith JD, Feagin JE |year=2011 |title=Branch point identification and sequence requirements for intron splicing in ''Plasmodium falciparum'' |journal=[[Eukaryotic Cell]] |doi=10.1128/EC.05193-11 |volume=10 |issue=11 |pages=1422}}</ref> The 5' and 3' splice sites agree with the [[canonical sequence]]s (GT and AG respectively). The 5' consensus motif is weakly conserved and tolerates nucleotide substitution including the fifth nucleotide in the intron. This fifth position, typically a G nucleotide in most eukaryotes, is frequently an A in ''P. falciparum''. The 3' splice site has a strong eukaryotic consensus sequence and a conserved adjacent polypyrimidine tract. The branch point is less well conserved with multiple branch points per intron with some at U instead of the typical A residue. A weak branch point consensus has been identified. |
|||
==Proteome== |
|||
There are 5,268 predicted proteins in ''[[Plasmodium falciparum]]'' and roughly 60% share little or no similarity to proteins in other organisms and thus are without functional assignment.<ref name="gardner"/> Of the predicted proteins, 31% contain at least one transmembrane domain and 17.3% have a signal peptide or signal anchor.<ref name="gardner"/> |
|||
It is estimated that 10.4% of the [[proteome]] is targeted to the [[apicoplast]] and 4.7% to the mitochondria.<ref name="gardner"/> |
|||
The parasite has different subsets of its proteome expressed during various stages of its developmental cycle.<ref name="florens">{{cite journal |
|||
|title=A proteomic view of the ''Plasmodium falciparum'' life cycle |
|||
|journal=Nature |
|||
|volume=419 |
|||
|issue=6906 |
|||
|pages=520–526 |
|||
|date=3 October 2002 |
|||
|doi=10.1038/nature01107 |
|||
|pmid=12368866 |
|||
|last1=Florens |
|||
|first1=Laurence |
|||
|last2=Washburn |
|||
|first2=Michael P. |
|||
|last3=Raine |
|||
|first3=J. Dale |
|||
|last4=Anthony |
|||
|first4=Robert M. |
|||
|last5=Grainger |
|||
|first5=Munira |
|||
|last6=Haynes |
|||
|first6=J. David |
|||
|last7=Moch |
|||
|first7=J. Kathleen |
|||
|last8=Muster |
|||
|first8=Nemone |
|||
|last9=Sacci |
|||
|first9=John B. |
|||
|last10=Tabb |
|||
|first10=David L. |
|||
|last11=Witney |
|||
|first11=Adam A. |
|||
|last12=Wolters |
|||
|first12=Dirk |
|||
|last13=Wu |
|||
|first13=Yimin |
|||
|last14=Gardner |
|||
|first14=Malcolm J. |
|||
|last15=Holder |
|||
|first15=Anthony A. |
|||
|last16=Sinden |
|||
|first16=Robert E. |
|||
|last17=Yates |
|||
|first17=John R. |
|||
|last18=Carucci |
|||
|first18=Daniel J.}}</ref> In one study, of the 2,415 proteins were identified in four stages(sporozoite, merozoite, trophozoite, gametocyte), representing 46% of the theoretical number of proteins.<ref name="florens"/> Only 6% of the proteins were found in all of the four stages. Of the proteins found, 51% were annotated as hypothetical proteins. |
|||
[[Merozoites]] contained high levels of cell recognition and invasion proteins. [[Trophozoites]] contained proteins implicated in [[erythrocyte]] remodeling and [[hemoglobin]] digestion. Gametocytes contained high amounts of gametocyte-specific [[transcription factors]] and cell cycle/DNA processing proteins. The gametocytes had low levels of polymorphic surface antigens. Sporozoites contained large amounts of proteins related to invasion, as well as members of the ''var'' and ''rif'' families.<ref name="florens"/> |
|||
===Translation initiation=== |
|||
This has been examined experimentally for the heat shock protein 86.<ref name=Patakottu2011>{{cite journal |author=Patakottu BR, Singh PK, Malhotra P, Chauhan VS, Patankar S |year=2011 |title=''In vivo'' analysis of translation initiation sites in ''Plasmodium falciparum'' |journal=Mol Biol Rep}}</ref> Like other eukaryotes purines at the -3 and +4 positions are essential for efficient translation. [[Uracil]] at the -1 position resulted in 2.5-fold higher reporter activity compared to wild type. |
|||
==Mosquito bite== |
|||
''[[Plasmodium falciparum|P. falciparum]]'' is transmitted to humans by the females of the ''[[Anopheles]]'' species of [[mosquito]]. There are about 460 species of ''[[Anopheles]]'' [[mosquito]], but only 68 transmit malaria. ''[[Anopheles gambiae]]'', found in Africa, is one of the best malaria vectors. It is long-lived, prefers feeding on humans, and lives in areas near human habitation.<ref name="1.3">{{cite web |
|||
|title=Malaria eModule - Transmission |
|||
|url=http://www.impact-malaria.com/iml/cx/en/translatedcontent.jsp?tcnt=_&tscat=_&tulg=CX_EN}}{{dead link|date=October 2013}}</ref> |
|||
Prior to transmission, ''[[Plasmodium falciparum]]'' resides within the salivary gland of the [[mosquito]]. The parasite is in its [[sporozoite]] stage at this point. The Pumilio-FBF family member Puf2 appears to be critical for its survival in the mosquito salivary gland.<ref name=Lindner2013>Lindner SE, Mikolajczak SA, Vaughan AM, Moon W, Joyce BR, Sullivan WJ Jr, Kappe SH (2013) Perturbations of ''Plasmodium'' Puf2 expression and RNA-seq of Puf2-Deficient sporozoites reveal a critical role in maintaining RNA homeostasis and parasite transmissibility. ''Cell Microbiol.'' 2013 Jan 29. {{DOI|10.1111/cmi.12116}}</ref> |
|||
As the [[mosquito]] takes its blood meal, it injects a small amount of saliva into the skin wound. The saliva contains antihemostatic and anti-inflammatory enzymes that disrupt the clotting process and inhibit the pain reaction.<ref name="ano-ms">{{cite web |
|||
|title=Malaria Site: Anopheles Mosquito |
|||
|url=http://www.malariasite.com/malaria/AnophelesMosquito.htm}}</ref> Some of the details of this process are known. A salivary protein - [[anophelin]] - is a powerful [[thrombin]] inhibitor. This protein occupies thrombin's active site but is highly resistant to cleavage by the enzyme.<ref name=Figueiredo2012>Figueiredo AC, de Sanctis D, Gutiérrez-Gallego R, Cereija TB, Macedo-Ribeiro S, Fuentes-Prior P, Pereira PJ (2012) Unique thrombin inhibition mechanism by anophelin, an anticoagulant from the malaria vector. Proc Natl Acad Sci USA</ref> |
|||
Typically, each infected bite contains 5-200 [[sporozoites]] which proceed to infect the human vector.<ref name="1.3"/> Once in the human bloodstream, the sporozoites only circulate for a matter of minutes before infecting liver cells. |
|||
~1900 proteins have been reported from sporozoites infecting the salivary glands.<ref name=Lindner2013>Lindner SE, Swearingen KE, Harupa A, Vaughan AM, Sinnis P, Moritz RL, Kappe SH (2013) Total and putative surface proteomics of malaria parasite salivary gland sporozoites. Mol Cell Proteomics</ref> |
|||
==Liver stage== |
|||
After circulating in the bloodstream, the ''[[Plasmodium falciparum|P. falciparum]]'' [[sporozoites]] enter [[hepatocytes]]. Before entering a hepatocyte the sporozoite typically engages in traversal of several cells. The reason for this behavior is not clear but it appears to reduce clearance of the sporozoites by [[Kupffer cell]]s.<ref name=Tavares2013>Tavares J, Formaglio P, Thiberge S, Mordelet E, Van Rooijen N, Medvinsky A, Ménard R, Amino R (2013) Role of host cell traversal by the malaria sporozoite during liver infection. J Exp Med</ref> |
|||
Once within a hepatocyte the parasite loses its [[apical complex]] and surface coat and transforms into a [[trophozoite]]. Within the parasitophorous vacuole of the [[hepatocyte]], ''[[Plasmodium falciparum|P. falciparum]]'' undergoes schizogonic development. In this stage, the nucleus divides multiple times with a concomitant increase in cell size but without cell segmentation. This exoerythrocytic schizogony stage of ''P. falciparum'' has a minimum duration of roughly 5.5 days. After segmentation, the parasite cells differentiate into [[merozoites]].<ref name="1.4">{{cite web |
|||
|title=Malaria eModule - Exo-Erythrocytic Stages |
|||
|url=http://www.impact-malaria.com/FR/EPS/Formations_et_cours_internationaux/Formation_de_la_Liverpool_School_LSTMH/cours_liverpool/Unit_1/1_4.html}}{{dead link|date=October 2013}}</ref> |
|||
Invasion of the hepatocytes appears to involve at least 2 proteins: sporozoite invasion-associated proteins (SIAP)-1 and -2.<ref name="Arévalo-Pinzón2011">{{cite journal |author=Arévalo-Pinzón G, Curtidor H, Muñoz M, Patarroyo MA, Patarroyo ME |year=2011 |title=Synthetic peptides from two Pf sporozoite invasion-associated proteins specifically interact with HeLa and HepG2 cells |journal=Peptides |pmid=21864602 |doi=10.1016/j.peptides.2011.08.008 |volume=32 |issue=9 |pages=1902–8}}</ref> These proteins bind [[heparin]] sulfate and [[chondroitin]] sulfate type membrane receptors on host cells. |
|||
Productive invasion of the hepatocyte results in the creation of a digestive vacuole, while merely passing through to reach another hepatocyte does not. Invasion of the liver cell changes the properties of the cell itself. The cell membrane becomes rougher and the cell itself becomes significantly stiffer.<ref name=Eaton2011>{{cite journal |author=Eaton P, Zuzarte-Luis V, Mota MM, Santos NC, Prudêncio M |year=2011 |title=Infection by ''Plasmodium'' changes shape and stiffness of hepatic cells |journal=Nanomedicine |pmid=22033078 |doi=10.1016/j.nano.2011.10.004 |volume=8 |issue=1 |pages=17–9}}</ref> The mechanism of these changes is currently unknown. |
|||
During this stage of development the sporozoite selectively discards organelles unnecessary for growth at this stage of the life cycle. Among these are the micronemes and the inner membrane complex.<ref name=Coppens2011>{{cite journal |author=Coppens I |title=Metamorphoses of malaria: the role of autophagy in parasite differentiation |journal=Essays Biochem. |volume=51 |pages=127–36 |year=2011 |pmid=22023446 |doi=10.1042/bse0510127}}</ref> |
|||
The division of the liver stages into thousands of merozoites is a complex process. In parallel with nuclear division, the apicoplast and mitochondrion become two extensively branched and intertwining structures.<ref name=Stanway2011>{{cite journal |author=Stanway RR, Mueller N, Zobiak B, ''et al''. |title=Organelle segregation into Plasmodium liver stage merozoites |journal=Cell. Microbiol. |volume=13 |issue=11 |pages=1768–82 |year=2011 |month=November |pmid=21801293 |doi=10.1111/j.1462-5822.2011.01657.x}}</ref> The organelles subsequently undergo morphological and positional changes prior to cell division. Finally to form merozoites, the parasite undergoes cytokinesis. |
|||
After maturation, the [[merozoites]] are released from the [[hepatocytes]] and enter the erythrocytic portion of their life-cycle. Note that these cells do not reinfect [[hepatocytes]]. |
|||
===Molecular biology of liver stages=== |
|||
''Plasmodium'' possess only a single [[pyruvate dehydrogenase]] enzyme (PDH) complex. This is localized to the plastid-like organelle known as the [[apicoplast]]. Unlike most eukaryotes, ''Plasmodium'' lacks a [[mitochondria]]l PDH. The PDH catalyzes the conversion of pyruvate to [[acetyl-CoA]], an important precursor for the [[tricarboxylic acid cycle]] and type II [[fatty acid]] synthesis. |
|||
The process of maturation within the liver is still being investigated. In the species ''[[Plasmodium yoelli]]'' the exit from the liver appears to be dependent on type II fatty acid synthesis.<ref>{{cite journal |author=Pei Y, Tarun AS, Vaughan AM, Herman RW, Soliman JM, Erickson-Wayman A, Kappe SH. |year=2010 |title=''Plasmodium'' pyruvate dehydrogenase activity is only essential for the parasite's progression from liver infection to blood infection |journal=Microbiol. |pmid=20487290 |doi=10.1111/j.1365-2958.2009.07034.x |volume=75 |issue=4 |pages=957–71}}</ref> Deletions in either the pyruvate dehydrogenase E1 alpha and E3 subunits produce a phenotype similar to that found in mutants of the type II fatty acid synthesis pathway. These mutants appear normal in blood stage development, mosquito stage development and early liver stage development but fail to exit the liver cells. |
|||
''Plasmodium'' is unable to synthesize [[sterol]]s they must obtain these from the host. However manipulation of cholesterol metabolism does not impede the development of the merozoites.<ref>{{cite journal |author=Labaied M, Jayabalasingham B, Bano N, Cha SJ, Sandoval J, Guan G, Coppens I |year=2010 |title=''Plasmodium'' salvages cholesterol internalized by LDL and synthesized de novo in the liver |journal=Cell Microbiol. |doi=10.1111/j.1462-5822.2010.01555.x |volume=13 |issue=4 |pages=569–86 |pmid=21105984}}</ref> |
|||
Invasion of the hepatocyte induces the production of [[CD81]] - a member of the tetraspanin superfamily.<ref name=Duarte2012>Duarte J, Herbert F, Guiyedi V, Franetich JF, Roland J, Cazenave PA, Mazier D, Kombila M, Fesel C, Pied S (2012) High levels of IgE autoantibody to 14-3-3 epsilon protein correlate with protection against severe ''Plasmodium falciparum'' malaria. J Infect Dis</ref> CD81 also appears to play a role in [[liver]] invasion by ''[[Plasmodium]]'' species.<ref name="pmid18389082">{{cite journal |author=Yalaoui S, Zougbédé S, Charrin S, Silvie O, Arduise C, Farhati K, Boucheix C, Mazier D, Rubinstein E, Froissard P |title=Hepatocyte permissiveness to ''Plasmodium'' infection is conveyed by a short and structurally conserved region of the CD81 large extracellular domain |journal=PLoS Pathogens |volume=4 |issue=2 |pages=e1000010 |year=2008 |month=February |pmid=18389082 |pmc=2279262 |doi=10.1371/journal.ppat.1000010 |editor1-last=Mota |editor1-first=Maria M}}</ref> It is required for ''[[Plasmodium vivax]]'' sporozoite entry into human hepatocytes and for ''[[Plasmodium yoelii]]'' sporozoite entry into murine hepatocytes.<ref name="pmid12483205">{{cite journal |author=Silvie O, Rubinstein E, Franetich JF, Prenant M, Belnoue E, Rénia L, Hannoun L, Eling W, Levy S, Boucheix C, Mazier D |title=Hepatocyte CD81 is required for ''Plasmodium falciparum'' and ''Plasmodium yoelii'' sporozoite infectivity |journal=Nat. Med. |volume=9 |issue=1 |pages=93–6 |year=2003 |month=January |pmid=12483205 |doi=10.1038/nm808}}</ref> |
|||
The protein UIS3 is an essential protein for liver stage development.<ref name=Favretto2012>Favretto F, Assfalg M, Molinari H, D'Onofrio M (2012) Evidence from NMR interaction studies challenges the hypothesis of direct lipid transfer from L-FABP to malaria sporozoite protein UIS3. Protein Sci. 2012 Nov 20. {{DOI|10.1002/pro.2194}}</ref> It is thought to be localised to the membrane of the parasitophorous vacuole of the infected cell. |
|||
Latency of sporozoites is controlled by the [[eIF2]] alpha kinase [[IK2]], a general inhibitor of protein synthesis.<ref name="Müller2011">{{cite journal |author=Müller K, Matuschewski K, Silvie O |year=2011 |title=The Puf-family RNA-binding protein Puf2 controls sporozoite conversion to liver stages in the malaria parasite |journal=PLoS ONE |volume=6 |issue=5 |page=e19860 |doi=10.1371/journal.pone.0019860 |editor1-last=Gruner |editor1-first=Anne Charlotte |pmid=21673790 |pmc=3097211}}</ref> Puf2 participates in the regulation of IK2 and inhibits premature sporozoite transformation. In contrast Puf1 appears to be dispensable. |
|||
The RNA binding protein family PUF member [[Pumilio]]-2 (Puf2) appears to be involved in transformation of sporozoites into the hepatic stage of the life cycle.<ref name=Gomes-Santos2011>{{cite journal |author=Gomes-Santos CS |year=2011 |title=Transition of ''Plasmodium'' sporozoites into liver stage-like forms is regulated by the RNA binding protein pumilio |journal=PLoS Pathog |volume=7 |issue=5 |page=e1002046 |author-separator=, |author2=Braks J |author3=Prudêncio M |author4=Carret C |author5=Gomes AR |author6=Pain A |author7=Feltwell T |author8=Khan S |author9=Waters A |display-authors=9 |pmc=3098293 |pmid=21625527 |doi=10.1371/journal.ppat.1002046 |editor1-last=Soldati-Favre |editor1-first=Dominique |first10=Chris |first11=Gunnar R.}}</ref> Knock out mutants of this gene exhibit genome wide transcriptional changes resulting in loss of gliding motility, cell traversal ability, reduction in infectivity and trigger metamorphosis typical of early ''Plasmodium'' intra-hepatic development. |
|||
Type II [[fatty acid]] biosynthesis is vital for this stage in the life cycle.<ref name=Yu2008>{{cite journal |author=Yu M, Kumar T. R, Nkrumah L. J, Coppi A, Retzlaff S ''et al.'' |year=2008 |title=The fatty acid biosynthesis enzyme FabI plays a key role in the development of liver-stage malarial parasites |journal=Cell Host & Microbe |volume=4 |issue=6 |pages=567–578 |pmid=19064257 |doi=10.1016/j.chom.2008.11.001 |pmc=2646117}}</ref> This pathway may be inhibited by the antibiotic [[triclosan]]. |
|||
The host iron regulatory hormone [[hepcidin]] which is synthesised in the liver and [[spleen]], appears to be able to inhibit growth of the liver stages.<ref name=Portugal2011>{{cite journal |author=Portugal S |year=2011 |title=Host-mediated regulation of superinfection in malaria |journal=Nat Med |volume=17 |issue=6 |pages=732–737 |author-separator=, |author2=Carret C |author3=Recker M |author4=Armitage AE |author5=Gonçalves LA |author6=Epiphanio S |author7=Sullivan D |author8=Roy C |author9=Newbold CI |display-authors=9 |doi=10.1038/nm.2368 |pmid=21572427 |first10=Hal |first11=Maria M}}</ref> Levels of this hormone are elevated during infection and seem to correlate with the anaemia often found in malaria.<ref name=deMast2009>{{cite journal |author=de Mast Q, Nadjm B, Reyburn H, ''et al''. |title=Assessment of urinary concentrations of hepcidin provides novel insight into disturbances in iron homeostasis during malarial infection |journal=J. Infect. Dis. |volume=199 |issue=2 |pages=253–62 |year=2009 |month=January |pmid=19032104 |doi=10.1086/595790}}</ref> Erythrocytic parasitaemia, above a minimum threshold, impairs the growth of subsequent liver cell sporozoite infection.<ref name=Portugal2011>Portugal S, Drakesmith H, Mota MM (2011) Superinfection in malaria: ''Plasmodium'' shows its iron will. EMBO Rep. {{doi|10.1038/embor.2011.213}}</ref> The production of hepcidin leads to the redistributes iron away from hepatocytes thus slowing the development of the iron dependent liver stage. |
|||
Liver hepcidin expression is upregulated and downregulated during the early and late stages of malaria infection respectively.<ref name=Wang2011>{{cite journal |author=Wang HZ, He YX, Yang CJ, Zhou W, Zou CG |year=2011 |title=Hepcidin is regulated during blood-stage malaria and plays a protective role in malaria infection |journal=J Immunol |pmid=22084434 |doi=10.4049/jimmunol.1101436 |volume=187 |issue=12 |pages=6410–6}}</ref> Inflammation and erythropoietin, rather than the [[iron]] sensing pathway, are involved in the regulation of hepcidin expression. Treatment of malaria infected mice with anti hepcidin neutralizing antibodies increased parasitemia and mortality rates. Overexpression of hepcidin improves the outcome. |
|||
[[Lipocalin]] 2, a host protein that sequesters iron, is upregulated during infection and appears to be involved in the host response.<ref name=Zhao2012>Zhao H, Konishi A, Fujita Y, Yagi M, Ohata K, Aoshi T, Itagaki S, Sato S, Narita H, Abdelgelil NH, Inoue M, Culleton R, Kaneko O, Nakagawa A, Horii T, Akira S, Ishii KJ, Coban C (2012) Lipocalin 2 bolsters innate and adaptive immune responses to blood-stage malaria infection by reinforcing host iron metabolism. Cell Host Microbe 12(5) 705-16. {{DOI|10.1016/j.chom.2012.10.010}}</ref> This protein increases both host [[macrophage]] function and [[granulocyte]] recruitment and decreases [[reticulocytosis]]. |
|||
Expression of the iron sequestering protein [[ferritin]] (ferritin H chain in mice) is associated with decreased tissue damage.<ref name=Gozzelino2012>Gozzelino R, Andrade BB, Larsen R, Luz NF, Vanoaica L, Seixas E, Coutinho A, Cardoso S, Rebelo S, Poli M, Barral-Netto M, Darshan D, Kühn LC, Soares MP (2012) Metabolic adaptation to tissue iron overload confers tolerance to malaria. ''Cell Host Microbe'' 12(5) 693-704. {{DOI|10.1016/j.chom.2012.10.011}}</ref> The mechanism appears to be via prevention of activation of the proapoptotic c-Jun N-terminal kinase. |
|||
Invasion of the hepatocyte seems to require the RON4 protease.<ref name=Giovannini2011>{{cite journal |author=Giovannini D, Späth S, Lacroix C, ''et al''. |title=Independent roles of apical membrane antigen 1 and rhoptry neck proteins during host cell invasion by apicomplexa |journal=Cell Host Microbe |volume=10 |issue=6 |pages=591–602 |year=2011 |month=December |pmid=22177563 |doi=10.1016/j.chom.2011.10.012}}</ref> |
|||
Within the liver [[actin]] reorganization is a dynamic process in part controlled by the actin severing and capping protein - [[gelsolin]].<ref name=Gomes-Santos2012>{{cite journal |author=Gomes-Santos CS, Itoe MA, Afonso C, ''et al''. |title=Highly Dynamic Host Actin Reorganization around Developing Plasmodium Inside Hepatocytes |journal=PLoS ONE |volume=7 |issue=1 |pages=e29408 |year=2012 |pmid=22238609 |pmc=3253080 |doi=10.1371/journal.pone.0029408 |editor1-last=Kappe |editor1-first=Stefan}}</ref> The hepatocyte cytoskeleton may contribute to parasite elimination. |
|||
Within the genome is encoded a homolog of [[macrophage migration inhibitory factor]]. This gene appears to be important for parasite development in the liver.<ref name=Miller2012>{{cite journal |author=Miller JL, Harupa A, Kappe SH, Mikolajczak SA |year=2012 |title=''Plasmodium'' macrophage migration inhibitory factor is necessary for efficient liver stage development |doi=10.1128/IAI.05861-11 |journal=Infect Immun |volume=80 |issue=4 |pages=1399–407 |pmid=22252874 |pmc=3318411}}</ref> |
|||
In ''Plasmodium bergei'' a protein - liver specific protein 2 (LISP2) - is expressed 24 hours after infection and rapidly increases during the liver stage schizogony. LISP2 is carried first to the parasitophorous vacuole and subsequently transported to the cytoplasm and nucleus of host hepatocytes. Mutations in this gene result in arrested development of the merozoites.<ref>Orito Y, Ishino T, Iwanaga S, Kaneko I, Kato T, Menard R, Chinzei Y, Yuda M (2012) Liver-specific protein 2: a ''Plasmodium'' protein exported to the hepatocyte cytoplasm and required for merozoite formation. Mol Microbiol {{DOI|10.1111/mmi.12083}}</ref> |
|||
Prior to transmission, ''[[Plasmodium falciparum]]'' resides within the salivary glad of the [[mosquito]]. The parasite is in its [[sporozoite]] stage at this point. As the [[mosquito]] takes its blood meal, it injects a small amount of saliva into the skin wound. The saliva contains antihemostatic and anti-inflammatory enzymes that disrupt the clotting process and inhibit the pain reaction.<ref name="ano-ms">{{cite web |
|||
| last = |
|||
| first = |
|||
| authorlink = |
|||
| coauthors = |
|||
| title = Malaria Site: Anopheles Mosquito |
|||
| work = |
|||
| publisher = |
|||
| date = |
|||
| url = http://www.malariasite.com/malaria/AnophelesMosquito.htm |
|||
| format = |
|||
| doi = |
|||
| accessdate = }}</ref> Typically, each infected bite contains 5-200 [[sporozoite]]s which proceed to infect the human vector.<ref name="1.3"/> Once in the human bloodstream, the [[sporozoite]]s only circulate for a matter of minutes before infecting liver cells. |
|||
Two other proteins (p52 and p36)in ''Plasmodium bergei'' appear to be important in the formation of the parasitophorous vacuole membrane in the liver.<ref name=Ploemen2012>Ploemen IH, Croes HJ, van Gemert GJ, Wijers-Rouw M, Hermsen CC, Sauerwein RW (2012) ''Plasmodium berghei'' Δp52&p36 Parasites develop independent of a parasitophorous vacuole membrane in Huh-7 liver cells" ''PLoS One'' 7(12) e50772. {{DOI|10.1371/journal.pone.0050772}}</ref> |
|||
===Liver stage=== |
|||
After circulating in the bloodstream, the ''[[Plasmodium falciparum|P. falciparum]]'' [[sporozoites]] enter [[hepatocytes]]. At this point, the parasite loses its [[apical complex]] and surface coat, and transforms into a [[trophozoite]]. Within the parasitophorous vacuole of the [[hepatocyte]], ''[[Plasmodium falciparum|P. falciparum]]'' undergoes schizogonic development. In this stage, nucleus divides multiple times with a concomitant increase in cell size, but without cell segmentation. This exoerythrocytic schizogony stage of ''[[Plasmodium falciparum|P. falciparum]]'' has a minimum duration of roughly 5.5 days. After segmentation, the parasite cells are differentiated into [[merozoites]].<ref name="1.4">{{cite web |
|||
| last = |
|||
| first = |
|||
| authorlink = |
|||
| coauthors = |
|||
| title = Malaria eModule - Exo-Erythrocytic Stages |
|||
| work = |
|||
| publisher = |
|||
| date = |
|||
| url = http://www.impact-malaria.com/FR/EPS/Formations_et_cours_internationaux/Formation_de_la_Liverpool_School_LSTMH/cours_liverpool/Unit_1/1_4.html |
|||
| format = |
|||
| doi = |
|||
| accessdate = }}</ref> |
|||
Infection of the liver induces [[apotosis]] in some of the liver cells.<ref name=Kaushansky2013>Kaushansky A, Metzger PG, Douglass AN, Mikolajczak SA, Lakshmanan V, Kain HS, Kappe SH (2013) Malaria parasite liver stages render host hepatocytes susceptible to mitochondria-initiated apoptosis. Cell Death Dis 4:e762. {{DOI|10.1038/cddis.2013.286}}</ref> Blocking this apototic response seems to increase the number of parasites in the liver suggesting that this may be a host defense mechanism. |
|||
After maturation, the [[merozoites]] are released from the [[hepatocyte]]s and enter the erythrocytic portion of their life-cycle. Note that these cells do not reinfect [[hepatocyte]]s. |
|||
==Erythrocytic stage== |
|||
[[Image:IEcycle.PNG|thumb|Plasmodium erythrocytic cycle<ref name="bozdech"/>]] |
[[Image:IEcycle.PNG|thumb|Plasmodium erythrocytic cycle<ref name="bozdech"/>]] |
||
'''''Merozoite''''' |
|||
===Effects on erythrocyte=== |
|||
After release from the [[hepatocyte]]s, the [[merozoites]] enter the bloodstream prior to infecting red blood cells. At this point, the [[merozoites]] are roughly 1.5 μm in length and 1 μm in diameter, and use the apicomplexan invasion organelles ([[apical complex]], pellicle and surface coat) to recognize and enter the host erythrocyte. |
|||
Infection of the [[erythrocyte]] induces a series of changes in the host cell's membrane. These changes depend on the stage of infection and are due to a subset of parasite derived proteins that are exported across the parasic vaculole membrane into the host cell's cytosol where they interact with the host cell cytoskeleton or are exposed at the erythrocyte surface. |
|||
Among these changes are a loss of deformability, an increase in rigidity and a novel propensity to adhere to vascular endothelial cells and unparasitized erythrocytes. Several proteins including [[knob-associated histidine rich protein]], [[Plasmodium falciparum erythrocyte membrane protein 3]], [[mature parasite infected erythrocyte surface antigen]] and [[ring parasite infected erythrocyte surface antigen]] are known to bind to the cytoskeleton and increase the erythrocyte's rigidity. Others including the [[Plasmodium falciparum erythrocyte membrane protein 1]] - the product of the ''var'' gene - are located on the surface of the erythrocyte and cause and are responsible for its new tendency to adhere. |
|||
'''''Trophozoite''''' |
|||
Another induced change is an alteration in the [[zeta potential]] - an electrochemical property of cell surfaces that is determined by the net electrical charge of molecules exposed at the surface of cell membranes - of the cell. The normal zeta potential of the erythrocyte is -15.7 [[volt|millivolts]] (mV).<ref name=Tokumasu2012>Tokumasu F, Ostera GR, Amaratunga C, Fairhurst RM (2012) Modifications in erythrocyte membrane zeta potential by ''Plasmodium falciparum'' infection. Exp Parasitol</ref> Much of this potential appears to be contributed by the exposed [[sialic acid]] residues in the membrane: their removal results in zeta potential of -6.06 mV. Infection of the erythrocyte decreases the mean to -14.6 mV at the trophozoite stage. Again removal of sialic acid in the infected cells increased the potential to -4.64 mV. |
|||
After invading the erythrocyte, the parasite loses its specific invasion organelles ([[apical complex]] and surface coat) and de-differentiates into a round [[trophozoite]] located within a parasitophorous vacuole in the red blood cell cytoplasm. The young [[trophozoite]] (or "ring" stage, because of its morphology on stained blood films) grows substantially before undergoing schizogonic division. <ref name="1.5">{{cite web |
|||
| last = |
|||
| first = |
|||
| authorlink = |
|||
| coauthors = |
|||
| title = Malaria eModule - ASEXUAL ERYTHROCYTIC STAGES |
|||
| work = |
|||
| publisher = |
|||
| date = |
|||
| url = http://www.impact-malaria.com/FR/EPS/Formations_et_cours_internationaux/Formation_de_la_Liverpool_School_LSTMH/cours_liverpool/Unit_1/1_5.html |
|||
| format = |
|||
| doi = |
|||
| accessdate = }}</ref> |
|||
A further change is the appearance of knobs - localised swellings of the erythrocyte membrane. The number of knobs, their diameter and height vary between isolates. They are thought to be important in the pathogenesis by contributing to blockage of capillaries. A number of parasite proteins are associated with these knobs including erythrocyte membrane protein 1 (the ''var'' gene product) and knob-associated histadine rich protein. Knobs first appear ∼20 hour post invasion and increase in number to ∼35 hours post-invasion.<ref name=Quadt2012>Quadt KA, Barfod L, Andersen D, Bruun J, Gyan B, Hassenkam T, Ofori MF, Hviid L (2012) The density of knobs on ''Plasmodium falciparum''-infected erythrocytes depends on developmental age and varies among isolates" ''PLoS One'' 7(9) e45658. {{DOI|10.1371/journal.pone.0045658}}</ref> They are more common in ''ex vivo'' isolates than in culture maintained strains. |
|||
'''''Schizont''''' |
|||
Singlet [[oxygen]] is generated during the parasite's life cycle. Its production is least in the ring stages, maximal in the schizonts and intermediate in the trophozoites.<ref name=Butzloff2012>Butzloff S, Groves MR, Wrenger C, Müller IB (2012) Cytometric quantification of singlet oxygen in the human malaria parasite ''Plasmodium falciparum''. Cytometry A {{doi|10.1002/cyto.a.22081}}.</ref> Its production may be related to the formation of haemozoin. |
|||
At the schizont stage, the parasite replicates its DNA multiple times without cellular segmentation. These schizonts then undergo cellular segmentation and differentiation to form roughly 16-18 merozoite cells in the erythrocyte.<ref name="1.5"/> The merozoites burst from the red blood cell, and proceed to infect other erythrocytes. The parasite is in the bloodstream for roughly 60 seconds before it has entered another erythrocyte.<ref name="cowman">{{cite journal |
|||
| last = Cowman |
|||
| first = |
|||
| authorlink = |
|||
| coauthors = |
|||
| title = Invasion of Red Blood Cells by Malaria Parasites |
|||
| journal = Cell |
|||
| volume = 124 |
|||
| issue = |
|||
| pages = 755–766 |
|||
| publisher = |
|||
| location = |
|||
| date = 24 February 2006 |
|||
| doi =10.1016/j.cell.2006.02.006 |
|||
| id = |
|||
| accessdate = }}</ref> |
|||
In normal erythrocytes the proteins [[ankyrin]], [[band 3]], [[band 4.1]], [[glycophorin]] and [[spectrin]] are phosphorylated. After invasion by the merozoite these proteins become dephosphorylated.<ref name=Murray1989>Murray MC, Perkins ME (1989) Phosphorylation of erythrocyte membrane and cytoskeleton proteins in cells infected with ''Plasmodium falciparum''. ''Mol Biochem Parasitol'' 34(3) 229-236</ref> As the parasite matures selective phosphorylation of ankyrin, band 3 and spectrin occurs. |
|||
This infection cycle occurs in a highly synchronous fashion, with roughly all of the parasites throughout the blood in the same stage of development. This precise clocking mechanism has been shown to be dependent on the human host's own [[circadian rhythm]].<ref name="1.5.2">{{cite web |
|||
| last = |
|||
| first = |
|||
| authorlink = |
|||
| coauthors = |
|||
| title = Malaria eModule - SYNCHRONICITY |
|||
| work = |
|||
| publisher = |
|||
| date = |
|||
| url = http://www.impact-malaria.com/FR/EPS/Formations_et_cours_internationaux/Formation_de_la_Liverpool_School_LSTMH/cours_liverpool/Unit_1/1_5_2.html |
|||
| format = |
|||
| doi = |
|||
| accessdate = }}</ref> Specifically, human body temperature changes, as a result of the circadian rhythm, seem to play a role in the development of ''[[Plasmodium falciparum|P. falciparum]]'' within the erythrocytic stage. |
|||
Tyrosine phosphorylation of band 3 at the ring stage appears to be under the control of [[Syk kinase]].<ref name=Pantaleo2010>Pantaleo A, Ferru E, Carta F, Mannu F, Giribaldi G, Vono R, Lepedda AJ, Pippia P, Turrini F (2010) Analysis of changes in tyrosine and serine phosphorylation of red cell membrane proteins induced by ''P. falciparum'' growth" ''Proteomics'' 10(19) 3469-3479</ref> Phosphorylation of additional cytoskeletal, trans-membrane and membrane associated proteins occurs as the parasite matures. These include [[actin]], [[adducin]]s, [[band 4.2]] and [[catalase]]. During the late schizont stage widespread protein dephosphorylation occurs. The erythrocyte kinases may be involved in this process. |
|||
Within the red blood cell, the parasite metabolism depends greatly on the digestion of [[hemoglobin]]. |
|||
Aggregation of the erythrocytes is known to occur during infection. This effect can be caused by culture supernatant suggesting a soluble product is responsible. Part of this mechanism appears to be the externalization of phosphatidyl-serine residues in the erythrocyte membrane. [[Methaemoglobin]] has been identified as the main causative agent of this alteration.<ref name=Balaji2013>Balaji SN, Trivedi V (2013) Extracellular methemoglobin primes red blood cell aggregation in malaria: An ''in vitro'' mechanistic study. FEBS Lett pii: S0014-5793(13)00003-3. {{DOI|10.1016/j.febslet.2012.12.015}}</ref> Its mechanism of action appears to be via the generation of reactive oxygen species: this action may be reversed with the addition of antioxidants. |
|||
Infected erythrocytes are often sequestered in various human tissues or organs, such as the heart, liver and brain. This is caused by parasite-derived cell surface proteins being present on the red blood cell membrane, and it is these proteins that bind to receptors on human cells. Sequestration in the brain causes cerebral malaria, a very severe form of the disease, which increases the victim's likelihood of death. |
|||
[[Metalloproteinase]] 9 is released from human microvascular endothelium after contact with infected erythrocytes.<ref name=D>D'Alessandro S, Basilico N, Prato M (2013) Effects of ''Plasmodium falciparum''-infected erythrocytes on matrix metalloproteinase-9 regulation in human microvascular endothelial cells. Asian Pac J Trop Med 6(3) 195-199 {{DOI|10.1016/S1995-7645(13)60022-X}}</ref> |
|||
The parasite can also alter the morphology of the red blood cell, causing knobs on the erythrocyte membrane. |
|||
Significant changes occur in the erthrocyte's cytoskeleton during infection.<ref name=Shi2013>Shi H, Liu Z, Li A, Yin J, Chong AG, Tan KS, Zhang Y, Lim CT (2013) Life cycle-dependent cytoskeletal modifications in ''Plasmodium falciparum'' infected erythrocytes" ''PLoS One'' 8(4) e61170. {{DOI|10.1371/journal.pone.0061170}}</ref> Among these are the accumulation of spectrin around the knobs and a decrease elsewhere. |
|||
The change in the shape of the erythrocyte induced by the parasite depends on the parasite species.<ref name=Karimi2013>Karimi A, Navidbakhsh M, Motevalli Haghi A, Faghihi S (2013) An innovative shape equation to quantify the morphological characteristics of parasitized red blood cells by ''Plasmodium falciparum'' and ''Plasmodium vivax''. Proc Inst Mech Eng H 227(4) 428-37. {{DOI|10.1177/0954411912474611}}</ref> Infection with ''P falciparum'' induces an increase in the cell volume of 80% in the ring stage. The erythrocyte subsequently becomes spherical in the trophozoite stage and remains so in the schizont stage. In contrast the volume increase induced by ''P vivax'' is only 30% and the erythrocyte remains biconcave throughout the infective cycle. |
|||
===Merozoite=== |
|||
After release from the [[hepatocytes]], the [[merozoites]] enter the bloodstream prior to infecting red blood cells. At this point, the merozoites are roughly 1.5 [[µm]] in length and 1 µm in diameter and use the apicomplexan invasion organelles ([[apical complex]], pellicle and surface coat) to recognize and enter the host erythrocyte. |
|||
The apicoplast measures 0.5 µm × 0.15 µm in the merozoite and is anchored to a band of 2-3 subpellicular microtubules.<ref name=Hopkins1999>Hopkins J, Fowler R, Krishna S, Wilson I, Mitchell G, Bannister L (1999) The plastid in ''Plasmodium falciparum'' asexual blood stages: a three-dimensional ultrastructural analysis. Protista 150:283–295</ref> Within the merozoite the mitochondrion and the apicoplast are aligned asymmetrically along the same side as the microtubules. |
|||
There are up to 40 micronemes per merozoite shaped like longnecked bottles. They are ~160 [[nanometer]]s (nm) long and 65 nm at their widest diameter.<ref name=Bannister2003>Bannister LH, Hopkins JM, Dluzewski AR, Margos G, Williams IT, Blackman MJ, Kocken CH, Thomas AW, Mitchell GH (2003) ''Plasmodium falciparum'' apical membrane antigen 1 (PfAMA-1) is translocated within micronemes along subpellicular microtubules during merozoite development" ''J Cell Sci'' 116(18) 3825-3834</ref> On their external surfaces, they bear bristle like filaments, each 3-4 nm thick and 25 nm long. The micronemes are translocated from a single Golgi like cisterna near the nucleus along a band of two or three subpellicular microtubules to the merozoite apex where they dock with the rhoptry tips. |
|||
Unlike species in the genus ''[[Toxoplasma]]'' which have multiple [[rhoptry|rhopteries]], ''Plasmodium'' species typically only have two. These may be referred to in the literature as the 'paired organ'. Several proteins have been localised to the rhopteries including asparagine rich parasite protein encoded on chromosome 4.<ref name=Wickramarachchi2013>Wickramarachchi T, Devi YS, Mohmmed A, Chauhan VS (2013) Identification and characterization of a novel ''Plasmodium falciparum'' merozoite apical protein involved in erythrocyte binding and invasion" ''PLoS One'' 3(3) e1732. {{DOI|10.1371/journal.pone.0001732}}</ref> Another is the Apical rhoptry neck protein whose expression is confined to the schizont stage.<ref name=Hans2013>Hans N, Singh S, Jain SK, Chauhan VS (2013) Identification of novel rhoptry neck protein of ''Plasmodium falciparum''. Mol Biochem Parasitol pii: S0166-6851(13)00032-7. {{DOI|10.1016/j.molbiopara.2013.02.007}}</ref> |
|||
After release from the erythrocyte the merozoites of the rodent malaria parasite ''[[Plasmodium yoelii]]'' change their shape from flat elongated ovals to spherical bodies.<ref name=Yahata2012>Yahata K, Treeck M, Culleton R, Gilberger TW, Kaneko O (2012) Time-lapse imaging of red blood cell invasion by the rodent malaria parasite ''Plasmodium yoelii''" ''PLoS One'' 7(12) e50780. {{DOI|10.1371/journal.pone.0050780}}</ref> This process takes ~60 seconds. During this time the merozoites were able to attach to and deform the erythrocyte membrane but were not able to reorient and invade. This morphological change may be related to the secretion or activation of invasion related proteins. |
|||
The parasite first binds to the erythrocyte in a random orientation. It then reorients such that the apical complex is in proximity to the erythrocyte membrane. A tight junction is formed between the parasite and erythrocyte. As it enters the red blood cell, the parasite forms a parasitophorous vesicle, to allow for its development inside the [[erythrocyte]].<ref name="cowman"/> |
|||
Five separate invasion paths have been delineated.<ref name=Chenniappan2011>{{cite journal |author=Chenniappan K |year=2011 |title=Alternative pathways of erythrocyte invasion, parasite multiplication potential and severity of the clinical episode of ''P. falciparum'' malaria in the Peruvian Amazon |journal=Parasitol Res |pmid=21993880 |doi=10.1007/s00436-011-2663-2 |volume=110 |issue=2 |pages=1019}}</ref> The most common pathway is [[neuraminidase]] resistant, [[trypsin]] sensitive and [[chymotrypsin]] resistant invasion. Some parasites have a neuraminidase- and trypsin-sensitive phenotype indicating a dependence on the erythrocyte binding antigen 175/[[GYPA|glycophorin A]] pathway(s). Most isolates appear to be dependent on a trypsin sensitive pathway. |
|||
====Erythrocyte invasion==== |
|||
This is a complex and poorly understood process. The merozoite initially contacts the erythrocyte and rotates until the rhoptery containing part is adjacent to the erythrocyte membrane. A tight contact is then established and the parasite enters the erythrocyte. This happens within seconds making the invasion process difficult to analyse. In ''Plasmodium yoelii'' the [[serine type serine repeat antigen]] (SERAs) are non essential for blood stage development of the parasite but appear to be an important factor in enabling the parasite to fully utilize the whole age repertoire of circulating erythrocytes.<ref name=Huang2013>Huang X, Liew K, Natalang O, Siau A, Zhang N, Preiser PR (2013) The role of serine-type serine repeat antigen in ''Plasmodium yoelii'' blood stage development" ''PLoS One'' 8(4) e60723. {{DOI|10.1371/journal.pone.0060723}}</ref> |
|||
In culture invasion of the erythrocytes can be prevent with the use of [[heparin]].<ref name=Miao2013>Miao J, Wang Z, Liu M, Parker D, Li X, Chen X, Cui L (2013) ''Plasmodium falciparum'': Generation of pure gametocyte culture by heparin treatment. Exp Parasitol pii: S0014-4894(13)00248-8. {{DOI|10.1016/j.exppara.2013.09.010}}</ref> Heparin binds only at the apical tip of the merozoite surface and multiple heparin binding proteins are found preferentially in the apical organelles.<ref name=Kobayashi2013>Kobayashi K, Takano R, Takemae H, Sugi T, Ishiwa A, Gong H, Recuenco FC, Iwanaga T, Horimoto T, Akashi H, Kato K (2013) Analyses of interactions between heparin and the apical ssurface proteins of ''Plasmodium falciparum''. Sci Rep 3:3178. doi: 10.1038/srep03178 </ref> The Duffy and reticulocyte binding-like families bind to heparin with diverse affinities suggesting that heparin masks the apical surface of merozoites and blocks interaction with the erythrocyte membrane after initial attachment. |
|||
Another protein that is bound by heparin is the merozoite surface protein 1.<ref name=Boyle2010>Boyle MJ, Richards JS, Gilson PR, Chai W, Beeson JG (2010) Interactions with heparin-like molecules during erythrocyte invasion by ''Plasmodium falciparum'' merozoites. Blood 115(22):4559-4568. doi: 10.1182/blood-2009-09-243725 </ref> |
|||
262 [[open reading frame]]s show sharp induction of expression during late schizont stages. At least 28 of these are proteins that are known to be involved in the invasion process. The functions of over 190 of these remain unknown.<ref name=Bozdech2003>Bozdech Z, Llinas M, Pulliam BL, Wong ED, Zhu J, ''et al'' (2003) The transcriptome of the intraerythrocytic developmental cycle of ''Plasmodium falciparum''. PLoS Biol 1:E5</ref> |
|||
====Initial adhesion==== |
|||
The rhopties are an ancient organelle within this group of protozoa while the micronemes appear to be a more recent development. The rhopteries are involved in cell invasion in other species in this clade. It would appear that the micronemal proteins have become adapted to recognise a cell potentially suitable for invasion and that the rhoptry proteins facilitate the invasion itself. It is likely that there is overlap of these functions particularly in the invasion step and that other proteins are also involved. |
|||
Exposure of the merozoites to a low [[potassium]] medium - at a concentration similar to that found in blood - induces a rise in the merozoite's cytosolic calcium concentration.<ref name="Singh2010">{{cite journal |last1=Singh |first1=S |last2=Alam |first2=MM |last3=Pal-Bhowmick |first3=I |last4=Brzostowski |first4=JA |last5=Chitnis |first5=CE |author-separator=, |year=2010 |title=Distinct external signals trigger sequential release of apical organelles during erythrocyte invasion by malaria parasites |journal=PLoS Pathog |volume=6 |issue=2 |page=e1000746 |doi=10.1371/journal.ppat.1000746 |editor1-last=Blackman |editor1-first=Michael John}}</ref> The low potassium levels in the blood (3.5-5.0 millimoles per liter) activates a [[phospholipase C]] enzyme. |
|||
The [[calcium dependent protein kinase]] 1 appears to play a role in micronene discharge.<ref name=Bansal2012>Bansal A, Singh S, More KR, Hans D, Nangalia K, Yogavel M, Sharma A, Chitnis CE (2012) Characterization of ''Plasmodium falciparum'' calcium dependent protein kinase 1 (PfCDPK1) and its role in microneme secretion during erythrocyte invasion. J Biol Chem</ref> The drug [[purfalcamine]] is a specific inhibitor of this kinase and it also inhibits micronene discharge and erythrocyte invasion. These kinases typically have an N terminal kinase domain and C terminal calmodulin like domain with calcium binding EF hands. The N and C terminals are joined by a junction domain. The C terminal appears to interact with the junction domain in the process of binding calcium. |
|||
[[Calcineurin]] a Ca<sup>2+</sup> dependent protein phosphatase is also involved in this process.<ref name=Singh2013>Singh S, More KR, Chitnis CE (2013) Role of calcineurin and actin dynamics in regulated secretion of microneme proteins in ''Plasmodium falciparum'' merozoites during erythrocyte invasion. Cell Microbiol {{DOI|10.1111/cmi.12177}}</ref> The dephosphorylation and depolymerization of apical actin also seems to be involved in this process. |
|||
[[Phospholipase C]] initiates a pathway that leads to a rise in the intracellular calcium. This rise in calcium triggers secretion of microneme proteins including the 175 kD [[erythrocyte binding antigen]] and [[apical membrane antigen 1]] to the merozoite surface where they can bind to erythrocyte surface proteins including [[GYPA|glycophorin A]]. The interaction of EBA175 with glycophorin A, its receptor on erythrocytes, restores basal cytosolic calcium levels. This in turn triggers the release of rhoptry proteins including the thrombospondin related apical merozoite protein (TRAMP).<ref name=Siddiqui2013>Siddiqui FA, Dhawan S, Singh S, Singh B, Gupta P, Pandey A, Mohmmed A, Gaur D, Chitnis CE (2013) A thrombospondin structural repeat containing rhoptry protein from ''Plasmodium falciparum'' mediates erythrocyte invasion. Cell Microbiol {{DOI|10.1111/cmi.12118}}</ref> |
|||
GAP45 is phosphorylated in response to [[Phospholipase C]] and calcium signaling.<ref name=Thomas2012>{{cite journal |last1=Thomas |first1=DC |last2=Ahmed |first2=A |last3=Gilberger |first3=TW |last4=Sharma |first4=P |year=2012 |title=Regulation of ''Plasmodium falciparum'' Glideosome Associated Protein 45 (PfGAP45) Phosphorylation |journal=PLoS ONE |volume=7 |issue=4 |page=e35855 |doi=10.1371/journal.pone.0035855 |editor1-last=Blader |editor1-first=Ira}}</ref> It is phosphorylated by the ''P. falciparum'' kinases [[Protein kinase B]] and [[Calcium dependent protein kinase 1]], both of which are calcium dependent enzymes, at Serine89, Serine103 and Serine149. Phosphorylation of these sites is differentially regulated during parasite development. |
|||
Two families of proteins are known to be involved in the invasion process: the reticulocyte binding like homologues (PfRh or PfRBP) and erythrocyte binding like (EBL) proteins. The EBL family are principally located in the micronemes and the Reticulocyte binding Homolog (PfRH) family are principally located in the rhopteries. Ligands from the EBL family largely govern the sialic acid dependent pathways of invasion and the RH family ligands (except for RH1) mediate sialic acid independent invasion.<ref name=Ord2012>{{cite journal |author=Ord RL, Rodriguez M, Yamasaki T, Takeo S, Tsuboi T, Lobo CA |title=Targeting Sialic Acid Dependent and Independent Pathways of Invasion in ''Plasmodium falciparum'' |journal=PLoS ONE |volume=7 |issue=1 |pages=e30251 |year=2012 |pmid=22253925 |pmc=3257272 |doi=10.1371/journal.pone.0030251 |editor1-last=Templeton |editor1-first=Thomas J}}</ref> During the invasion process these ligands are localized at the apical tip of the merozoite and are able to bind erythrocytes. |
|||
;Microneme proteins |
|||
All known micronemal proteins are type I integral membrane proteins that contain a C-terminal transmembrane domain and a short cytoplasmic domain.<ref name=Cowman2006>Cowman AF, Crabb BS (2006) Invasion of red blood cells by malaria parasites. Cell 124:755–766</ref> |
|||
The EBL family of proteins includes EBA-165 (also known as PEBL), EBA-175 (also known as PfF2), EBA-181 (also known as JESEBL), EBA-140 (also known as BAEBL) and EBL-1. Whilst these parasite ligands function in merozoite invasion by binding to specific receptors on the erythrocyte, they appear also to have a central role in activation of the invasion process. Binding of EBA-175 to its receptor, [[GYPA|glycophorin A]], restores the basal cytosolic calcium levels after interaction of the merozoite with the erythrocyte and triggers the release of rhoptry proteins.<ref name=Singh2010>{{cite journal |author=Singh S, Alam MM, Pal-Bhowmick I, Brzostowski JA, Chitnis CE |year=2010 |title=Distinct external signals trigger sequential release of apical organelles during erythrocyte invasion by malaria parasites |doi=10.1371/journal.ppat.1000746 |journal=PLoS Pathog |volume=6 |issue=2 |page=e1000746 |editor1-last=Blackman |editor1-first=Michael John}}</ref> |
|||
These proteins have several domains. Region II which is responsible for ligand erythrocyte interaction during invasion, consists of two homologous F1 and F2 domains.<ref name=Rydzak2012>Rydzak J, Kryńska K, Suchanowska A, Kaczmarek R, Lukasiewicz J, Czerwiński M, Jaśkiewicz E (2012) Bacterially expressed truncated F2 domain of ''Plasmodium falciparum'' EBA-140 antigen can bind to human erythrocytes. Acta Biochim Pol</ref> |
|||
Several of the receptors for the proteins are known: [[GYPA|glycophorin]] A for EBA-175, [[GYPB|glycophorin B]] for EBL-1 and [[glycophorin C]] for EBA-140.<ref name=Sim1994>Sim BKL, Chitnis CE, Wasniowska K, Hadley TJ, Miller LH (1994) Receptor and ligand domains for invasion of erythrocytes by ''Plasmodium falciparum''. Science 264:1941–1944</ref><ref name=Mayer2009>Mayer DC ''et al'' (2009) Glycophorin B is the erythrocyte receptor of ''Plasmodium falciparum'' erythrocyte-binding ligand, EBL-1. Proc Natl Acad Sci USA 106:5348–5352</ref><ref name=Maier2003>Maier AG ''et al'' (2003) ''Plasmodium falciparum'' erythrocyte invasion through glycophorin C and selection for Gerbich negativity in human populations. Nat Med 9:87–92</ref><ref name=Li2012>{{cite journal |author=Li X, Marinkovic M, Russo C, McKnight CJ, Coetzer TL, Chishti AH |year=2012 |title=Identification of a specific region of ''Plasmodium falciparum'' EBL-1 that binds to host receptor glycophorin B and inhibits merozoite invasion in human red blood cells |journal=Mol Biochem Parasitol |pmid=22273481 |doi=10.1016/j.molbiopara.2012.01.002 |volume=183 |issue=1 |pages=23–31 |pmc=3307866}}</ref> |
|||
The ''ebl'' family share a common intron/exon structure suggesting a common origin.<ref name=Adams1992>Adams JH, Sim BK, Dolan SA, Fang X, Kaslow DC, Miller LH (1992) A family of erythrocyte binding proteins of malaria parasites Proc. Natl Acad Sci USA 89:7085-7089</ref> These proteins also share a common organisation into several regions. Of these regions II and VI are the cysteine-rich regions of the DBL-EBP extracellular domains that have numerous conserved cysteine and hydrophobic amino acid residues, suggesting a conserved, functionally important three-dimensional structure. Despite this there is little nucleotide identity between the proteins. |
|||
The EBL proteins have a Duffy like binding domain (DBL) unique to ''Plasmodium'' species. The crystal structure of the binding domain of EBA-175 has been solved.<ref name=Ambroggio2013>Ambroggio X, Jiang L, Aebig J, Obiakor H, Lukszo J, Narum DL (2013) The epitope of monoclonal antibodies blocking erythrocyte invasion by ''Plasmodium falciparum'' map to The dimerization and receptor glycan binding Sites of EBA-175" ''PLoS One'' 8(2) e56326. {{DOI|10.1371/journal.pone.0056326}}</ref> It consists of a dimer of the beta fingers of the F1 and F2 regions. |
|||
The crystal structure of EBA-140 has been solved.<ref name=Lin2012>Lin DH, Malpede BM, Batchelor JD, Tolia NH (2012) Crystal and solution structures of ''Plasmodium falciparum'' erythrocyte binding antigen 140 reveal determinants of receptor specificity during erythrocyte invasion. J Biol Chem</ref> The two domain binding region is present as a monomer. Both domains are required for binding to occur. Its electrostatic surface has a basic patch spanning both DBL domains that is important in the binding mechanism. |
|||
EBA-175 mediates adhesion to erythrocytes through binding of the Duffy binding like domains in its extracellular domain to Neu5Acα2-3Galactose displayed on the O-linked glycans of glycophorin A.<ref name=Wanaguru2013>Wanaguru MK, Crosnier C, Johnson S, Rayner JC, Wright GJ (2013) A biochemical analysis of the ''Plasmodium falciparum'' erythrocyte binding antigen-175 (EBA175) - glycophorin-A interaction: implications for vaccine design. J Biol Chem</ref>. |
|||
A member of the EBL family of proteins (MAEBL) has been shown to be present in ''[[Plasmodium gallinaceum]]''.<ref name=Martinez2012>Martinez C, Marzec T, Smith CD, Tell LA, Sehgal RN (2012) Identification and expression of maebl, an erythrocyte-binding gene, in ''Plasmodium gallinaceum''. Parasitol Res</ref> This protein is now known to be conserved in the primate, rodent and avian infecting species suggesting that it may play an important role in erythrocyte invasion. The duplicate extracellular binding domains of MAEBL are responsible for erythrocyte binding. MAEBL is a type I transmembrane protein with a carboxyl cysteine rich region. |
|||
A GPI-anchored micronemal antigen (GAMA) also appears to be essential in the process of erythrocyte invasion.<ref name=Arumugam2011>{{cite journal |author=Arumugam TU, Takeo S, Yamasaki T, Thonkukiatkul A, Miura K, Otsuki H, Zhou H, Long CA, Sattabongkot J, Thompson J, Wilson DW, Beeson JG, Healer J, Crabb BS, Cowman AF, Torii M, Tsuboi T |year=2011 |title=Discovery of GAMA, a ''Plasmodium falciparum'' merozoite micronemal protein, as a novel blood-stage vaccine candidate antigen |journal=Infect Immun |pmid=21896773 |doi=10.1128/IAI.05412-11 |pmc=3257921 |volume=79 |issue=11 |pages=4523–32}}</ref> |
|||
A microneme associated antigen (PfMA: PF3D7_0316000, PFC0700c) binds erythrocytes in a sialic acid independent, chymotrypsin and trypsin resistant manner.<ref name=Hans2013>Hans N, Singh S, Pandey AK, Reddy KS, Gaur D, Chauhan VS (2013) Identification and characterization of a novel ''Plasmodium falciparum'' adhesin involved in erythrocyte invasion" ''PLoS One'' 8(9) e74790. {{DOI|10.1371/journal.pone.0074790}}</ref> This gene is expressed only in the late blood stages. It is a 307 amino acid protein (~ 37.1 kiloDalton) that contains an N-terminal stretch of hydrophobic residues, a C-terminal single transmembrane domain and a short cytoplasmic tail. It is moderately (~40%) conserved between several ''Plasmodium'' species. |
|||
;Rhoptery proteins |
|||
The PfRh family consists of five proteins and a [[pseudogene]]: PfRh1, PfRh2a, PfRh2b, PfRh3, PfRh4 and PfRh5. PfRh3 is a transcribed pseudogene in all strains examined to date.<ref name=Taylor2011>{{cite journal |author=Taylor HM |year=2001 |title=''Plasmodium falciparum'' homologue of the genes for ''Plasmodium vivax'' and ''Plasmodium yoelii'' adhesive proteins, which is transcribed but not translated |journal=Infect Immun |volume=69 |issue=6 |pages=3635–3645 |author-separator=, |author2=Triglia T |author3=Thompson J |author4=Sajid M |author5=Fowler R |display-authors=5 |doi=10.1128/IAI.69.6.3635-3645.2001 |pmid=11349024 |last6=Wickham |first6=ME |last7=Cowman |first7=AF |last8=Holder |first8=AA |pmc=98354}}</ref> All the other members of this family bind to erythrocyes and antibodies to them inhibit invasion. |
|||
The Pfh1 protein binds a sialic acid containing erythrocyte receptor.<ref name=Rayner2001>Rayner JC, Vargas-Serrato E, Huber CS, Galinski MR, Barnwell JW (2001) A ''Plasmodium falciparum'' homologue of ''Plasmodium vivax'' reticulocyte binding protein (PvRBP1) defines a trypsin-resistant erythrocyte invasion pathway. J Exp Med 194:1571–1581</ref> |
|||
PfRh1 and PfRh2 are located in the neck of the rhopteries.<ref name=Gao2008>Gao X, Yeo KP, Aw SS, Kuss C, Iyer JK, Genesan S, Rajamanonmani R, Lescar J, Bozdech Z, Preiser PR (2008) Antibodies targeting the PfRH1 binding domain inhibit invasion of ''Plasmodium falciparum'' merozoites. PLoS Pathog 4:e1000104 doi:10.1371/journal.ppat.1000104</ref><ref name=Duraisingh2003>Duraisingh MT, Triglia T, Ralph SA, Rayner JC, Barnwell JW, McFadden GI, Cowman AF (2003) Phenotypic variation of ''Plasmodium falciparum'' merozoite proteins directs receptor targeting for invasion of human erythrocytes. EMBO J 22:1047–1057</ref> |
|||
The genes PfRh2a and PfRh2b encode large proteins of about 3200 amino acids in length. They differ only in the last 500 amino acids of the C terminal and have clearly arisen by a process of gene duplication and mutation. The 500 amino acid region includes an ectodomain, a transmembrane domain and a cytoplasmic domain. PfRh2b is essential for a well-defined invasion pathway while PfRh2a is not required or sufficient for this pathway. It has been shown that the reason for this difference lies in the cytoplasmic domain.<ref name="Dvorin2010">{{cite journal |author=Dvorin JD, Bei AK, Coleman BI, Duraisingh MT |year=2010 |title=Functional diversification between two related ''Plasmodium falciparum'' merozoite invasion ligands is determined by changes in the cytoplasmic domain |journal=Mol Microbiol. |pmid=20487292 |doi=10.1111/j.1365-2958.2009.07040.x |volume=75 |issue=4 |pages=990–1006 |pmc=3627358}}</ref> |
|||
Reticulocyte binding like protein homologue 2a (PfRH2a) is processed both in the schizont as well as during invasion resulting in proteins with different erythrocyte binding properties.<ref name=Gunalan2011>{{cite journal |author=Gunalan K, Gao X, Liew KJ, Preiser PR |year=2011 |title=Differences in erythrocyte receptor specificity of different parts of the ''Plasmodium falciparum'' reticulocyte binding protein homologue 2a |journal=Infect Immun |doi=10.1128/IAI.00201-11 |volume=79 |issue=8 |pages=3421–30 |pmid=21628513 |pmc=3147545}}</ref> It also moves from the rhoptry neck to the moving junction during merozoite invasion. PfRh2a undergoes a cleavage event in the transmembrane region during invasion consistent with activity of the membrane associated PfROM4 protease.<ref name=Triglia2011>{{cite journal |author=Triglia T |year=2011 |title=''Plasmodium falciparum'' merozoite invasion is inhibited by antibodies that target the PfRh2a and b binding domains |journal=PLoS Pathog |volume=7 |issue=6 |page=e1002075 |author-separator=, |author2=Chen L |author3=Lopaticki S |author4=Dekiwadia C |author5=Riglar DT |author6=Hodder AN |author7=Ralph SA |author8=Baum J |author9=Cowman AF |display-authors=9 |doi=10.1371/journal.ppat.1002075 |editor1-last=Kazura |editor1-first=James W}}</ref> Both PfRh2a and PfRh2b bind to red blood cells. The erythrocyte-binding domain lies within a 15 kDa region at the N-terminus of each protein. |
|||
PfRh2b appears to play an important - if not dominant - role in the binding and invasion process.<ref name=Baum2005>Baum J, Maier AG, Good RT, Simpson KM, Cowman AF (2005) Invasion by ''P. falciparum'' merozoites suggests a hierarchy of molecular interactions" ''PLoS Pathog'' 1(4) e37</ref> |
|||
PfRh4 binds to the complement receptor 1 (CR1; [[CD35]]).<ref name=Park2013>Park HJ, Guariento M, Maciejewski M, Hauhart R, Tham WH, Cowman AF, Schmidt CQ, Mertens HD, Liszewski MK, Hourcade DE, Barlow PN, Atkinson JP (2013) Using mutagenesis and structural biology to map the binding site for the ''Plasmodium falciparum'' merozoite protein PfRh4 on the human immune adherence receptor. J Biol Chem </ref> Complement receptor 1 is a ∼190- to 280-kDa single-chain transmembrane glycoprotein and carries the Knops blood group antigen. The binding site lies within the three N-terminal complement control protein modules (CCPs 1-3) of CR1. This region also accommodate binding and regulatory sites for the key [[complement]] activation specific proteolytic products, C3b and C4b. The binding of Rh4 to CR1 does not inhibit the binding of C3b/C4b but it does inhibit their dissociation from the erythrocyte. The critical site for the binding of Rh4 appears to lie within the CCP-1 module. |
|||
PfRh4 is responsible for the majority (50–80%, depending on the parasite strain used) of the sialic acid independent invasion pathway.<ref name=Tham2009>Tham WH ''et al'' (2009) Antibodies to reticulocyte-binding protein-like homologue 4 inhibit invasion of ''Plasmodium falciparum'' into human erythrocytes. Infect Immun 77:2427–2435</ref> |
|||
PfRh4 binds to a second protein ''P. falciparum'' Rh5 interacting protein (PfRipr). PfRipr has a molecular weight of 123 kilo[[Dalton (unit)|Dalton]]s with 10 [[epidermal growth factor]]-like domains and 87 [[cysteine]] residues distributed along the protein.<ref name=Chen2011>{{cite journal |author=Chen L |year=2011 |title=An EGF-like protein forms a complex with PfRh5 and is required for invasion of human erythrocytes by ''Plasmodium falciparum'' |journal=PLoS Pathog |volume=7 |issue=9 |page=e1002199 |author-separator=, |author2=Lopaticki S |author3=Riglar DT |author4=Dekiwadia C |author5=Uboldi AD |author6=Tham WH |author7=O'Neill MT |author8=Richard D |author9=Baum J |display-authors=9 |pmid=21909261 |doi=10.1371/journal.ppat.1002199 |pmc=3164636 |editor1-last=Blackman |editor1-first=Mike John |first10=Stuart A. |first11=Alan F.}}</ref> |
|||
PfRh5 is located within the rhoptries and appears to be an essential gene.<ref name=Baum2009>{{cite journal |author=Baum J, Chen L, Healer J, ''et al''. |title=Reticulocyte-binding protein homologue 5 - an essential adhesin involved in invasion of human erythrocytes by ''Plasmodium falciparum'' |journal=Int. J. Parasitol. |volume=39 |issue=3 |pages=371–80 |year=2009 |month=February |pmid=19000690 |doi=10.1016/j.ijpara.2008.10.006}}</ref> |
|||
The receptor for the PfRh5 protein appears to be the Ok [[blood group antigen]], [[basigin]].<ref name=Crosnier2011>{{cite journal |author=Crosnier C, Bustamante LY, Bartholdson SJ, ''et al''. |title=Basigin is a receptor essential for erythrocyte invasion by ''Plasmodium falciparum'' |journal=Nature |volume=480 |issue=7378 |pages=534–7 |year=2011 |month=December |pmid=22080952 |pmc=3245779 |doi=10.1038/nature10606}}</ref> Blocking access to this protein on the erythrocyte surface appears to inhibit erythrocyte invasion completely. Binding of the Rh5 protein appears to be critically dependent on a single residue within the Rh5 protein.<ref name="Arévalo-Pinzón2011">Arévalo-Pinzón G, Curtidor H, Muñoz M, Patarroyo MA, Bermudez A, Patarroyo ME (2011) A single amino acid change in the ''Plasmodium falciparum'' RH5 (PfRH5) human RBC binding sequence modifies its structure and determines species-specific binding activity. Vaccine</ref> Antibodies to this protein inhibit the parasite's growth ''in vitro'' and appear to be correlated with protection against infection with it.<ref name=Tran2013>Tran TM, Ongoiba A, Coursen J, Crosnier C, Diouf A, Huang CY, Li S, Doumbo S, Doumtabe D, Kone Y, Bathily A, Dia S, Niangaly M, Dara C, Sangala J, Miller LH, Doumbo OK, Kayentao K, Long CA, Miura K, Wright GJ, Traore B, Crompton PD (2013) Naturally acquired antibodies specific for ''Plasmodium falciparum'' RH5 inhibit parasite growth and predict protection from malaria. J Infect Dis </ref> |
|||
There seems to be some overlap between the functions of these proteins. Loss of EBA-175 can be compensated by increased expression of PfRh4.<ref name=Lopaticki2011>{{cite journal |author=Lopaticki S |year=2011 |title=Reticulocyte and erythrocyte binding-like proteins function cooperatively in invasion of human erythrocytes by malaria parasites |journal=Infect Immun |volume=79 |issue=3 |pages=1107–1117 |author-separator=, |author2=Maier AG |author3=Thompson J |author4=Wilson DW |author5=Tham WH |display-authors=5 |doi=10.1128/IAI.01021-10 |pmid=21149582 |last6=Triglia |first6=T |last7=Gout |first7=A |last8=Speed |first8=TP |last9=Beeson |first9=JG |pmc=3067488 |first10=J. |first11=A. F.}}</ref> |
|||
The rhoptry associated, leucine zipper-like protein 1 (RALP1) is specifically expressed in schizont stages and localized to the rhoptry of merozoites. It appears to be an essential gene. It translocates from the rhoptry neck to the moving junction during merozoite invasion. Anti-RALP1 antibodies disrupt the tight junction formation. There is an erythrocyte binding domain in the C terminal. |
|||
Another protein localised to the apical ends of the rhoptries is an asparagine rich parasite protein (PfAARP;PFD1105w).<ref name=Wickramarachchi2008>Wickramarachchi T, Devi YS, Mohmmed A, Chauhan VS (2008) Identification and characterization of a novel ''Plasmodium falciparum'' merozoite apical protein involved in erythrocyte binding and invasion. PLoS One 3(3):e1732. doi: 10.1371/journal.pone.0001732</ref> This protein has a predicted signal sequence, a C-terminal transmembrane region and its transcription and translation patterns are similar to other merozoite surface proteins. |
|||
The rhoptry neck protein PfRON4 - a homologue of ''Toxoplasma gondii'' rhoptry neck protein TgRON4 - forms a complex with the protein PfAMA1 during its secretion in the course of merozoite invasion.<ref name=Alexander2006>Alexander DL, Arastu-Kapur S, Dubremetz JF, Boothroyd JC (2006) ''Plasmodium falciparum'' AMA1 binds a rhoptry neck protein homologous to TgRON4, a component of the moving junction in ''Toxoplasma gondii''. Eukaryot Cell 5:1169–1173.</ref> |
|||
Other rhoptery proteins include the rhoptry associated protein 1 (RAP1) and rhoptry associated membrane antigen (RAMA).<ref name=Clark1987>Clark JT, Anand R, Akoglu T, McBride JS (1987) Identification and characterisation of proteins associated with the rhoptry organelles of ''Plasmodium falciparum'' merozoites. Parasitol Res 73:425–434</ref><ref name=Topolska2004>Topolska AE, Lidgett A, Truman D, Fujioka H, Coppel RL (2004) Characterization of a membrane-associated rhoptry protein of ''Plasmodium falciparum''. J Biol Chem 279:4648–4656</ref> |
|||
The RhopH complex proteins localize to the basal bulb of the rhoptries and are involved in erythrocyte binding and in establishment of parasitophorous vacuole.<ref name=Ling2004>Ling IT, Florens L, Dluzewski AR, Kaneko O, Grainger M, ''et al'' (2004) The ''Plasmodium falciparum'' ''clag9'' gene encodes a rhoptry protein that is transferred to the host erythrocyte upon invasion. Mol Microbiol 52:107–118</ref><ref name=Hiller2003>Hiller NL, Akompong T, Morrow JS, Holder AA, Haldar K (2003) Identification of a stomatin orthologue in vacuoles induced in human erythrocytes by malaria parasites: A role for microbial raft proteins in Apicomplexan vacuole biogenesis. J Biol Chem 278:48413–48421</ref> |
|||
Apical membrane antigen 1 (AMA-1) is a type I transmembrane protein located in the neck of the malaria merozoite rhoptries and later on the surface of the invasive merozoite.<ref name=Peterson1989>Peterson MG, Marshall VM, Smythe JA, Crewther PE, Lew A, Silva A, Anders RF, Kemp DJ (1989) Integral membrane protein located in the apical complex of ''Plasmodium falciparum''. Mol Cell Biol 9:3151-3154</ref> Its ectodomain is defined by three cysteine-rich domains characterised by disulfide bond patterns. |
|||
;Merozoite surface protein family. |
|||
Merozoite surface protein 1 (MSP-1; P195; PMMSA; MSA 1) is a protein found on the surface of the merozoites.<ref name=Holder1992>Holder AA, Blackman MJ, Burghaus PA, Chappel JA, Ling IT, McCallum-Deighton N, Shai S (1992) A malaria merozoite surface protein (MSP1)-structure, processing and function. Mem Inst Oswaldo Cruz 87 Suppl 3:37-42</ref> During invasion of the new red cell most of the MSP1 molecule is shed from the parasite surface except for a small C-terminal fragment which can be detected in ring stages. Within this fragment are two [[epidermal growth factor]]-like domains. This protein has been found in all ''Plasmodium'' species studied to date suggesting it has an important role in the life cycle. |
|||
It appears that the merozoite surface protein 1 (MSP1) binds to [[heparin]] like molecules on the surface of the erythrocyte and that is binding is an essential step in the invasion process.<ref name="Boyle2010">{{cite journal |author=Boyle MJ, Richards JS, Gilson PR, Chai W, Beeson JG |year=2010 |title=Interactions with heparin-like molecules during erythrocyte invasion by ''P. falciparum'' merozoites |journal=Blood |pmid=20220119 |doi=10.1182/blood-2009-09-243725 |volume=115 |issue=22 |pages=4559–68}}</ref> |
|||
Merozoite surface protein 2 is one of the most abundant proteins on the surface of the merozoite and plays a role in the invasion process.<ref name=Zhang2013>Zhang X, Dong Y, Yu J, Tu X (2013) Effects of environmental factors on MSP21-25 aggregation indicate the roles of hydrophobic and electrostatic interactions in the aggregation process. Eur Biophys J </ref> Mediated by hdrophobic residues in the N terminal 1-25 residues it forms aggregates ''in vitro'' and may be present on the merozoite as aggregates. |
|||
In ''P. vivax'' there are a family (10) of merozoite surface protein 3 termed MSP3α to MSP3λ arranged in a head to tail fashion on chromosome 10.<ref name=Jiang2013>Jiang J, Barnwell JW, Meyer EV, Galinski MR. |
|||
''Plasmodium vivax'' merozoite surface protein-3 (PvMSP3) expression of an 11 member multigene family in blood-stage parasites" ''PLoS One'' 8(5) e63888. {{DOI|10.1371/journal.pone.0063888}}</ref> These proteins have a predominant central alanine rich domain containing heptad repeats predicted to form α-helical secondary and coiled-coil tertiary structures. They lack transmembrane domain or GPI-lipid modification site. |
|||
Merozoite surface protein 8 is a single open reading frame of 1791 base pairs which encodes a polypeptide of 597 amino acids.<ref name=Black2001>Black CG, Wu T, Wang L, Hibbs AR, Coppel RL (2001) Merozoite surface protein 8 of ''Plasmodium falciparum'' contains two epidermal growth factor-like domains. Mol Biochem Parasitol 114(2):217-226</ref> At the N-terminus there is a secretory signal peptide while at the and C-termini there is a GPI attachment sites. There are two EGF-like domains located near the C-terminus. |
|||
Merozoite surface protein 9 is conserved between species and appears to be under purifing selection.<ref name=Chenet2013>Chenet SM, Pacheco MA, Bacon DJ, Collins WE, Barnwell JW, Escalante AA (2013) The evolution and diversity of a low complexity vaccine candidate, merozoite surface protein 9 (MSP-9), in ''Plasmodium vivax'' and closely related species. Infect Genet Evol 20C:239-248. {{DOI|10.1016/j.meegid.2013.09.011}}</ref> |
|||
Merozoite surface proteins 8 and 10 which are thought to be involved in the invasion process appear to be under purifying selection.<ref name="Andreína2012">Andreína Pacheco M, Elango AP, Rahman AA, Fisher D, Collins WE, Barnwell JW, Escalante AA (2012) Evidence of purifying selection on merozoite surface protein 8 (MSP8) and 10 (MSP10) in ''Plasmodium'' spp. Infect Genet Evol</ref> |
|||
;Other proteins |
|||
Another family of proteins involved in the invasion process are the thrombospondin related anonymous protein (TRAP) family. These proteins are type I cell surface proteins with one or more extracellular thrombospondin type-I repeats (TSR) domains and/or von Willebrand factor like (vWF) A domain(s) and an acidic cytoplasmic tail with a subterminal tryptophan residue. The cytoplasmic tails of TRAP, CTRP, TLP and MTRAP interact with the enzyme aldolase. |
|||
One protein thought to be involved in the invasion process is the merozoite specific thrombospondin related anonymous protein homolog (MTRAP). The receptor for this protein has been identified as the GPI-linked protein [[SEMA7A|semaphorin-7A]] (CD108).<ref name=Bartholdson2012>Bartholdson SJ, Bustamante LY, Crosnier C, Johnson S, Lea S, Rayner JC, Wright GJ (2012) Semaphorin-7A is an erythrocyte receptor for ''P. falciparum'' merozoite-specific TRAP homolog, MTRAP" ''PLoS Pathog'' 8(11) e1003031. {{DOI|10.1371/journal.ppat.1003031}}</ref><ref name=Bartholdson2013>Bartholdson SJ, Crosnier C, Bustamante LY, Rayner JC, Wright GJ (2013) Identifying novel ''Plasmodium falciparum'' erythrocyte invasion receptors using systematic extracellular protein interaction screens. Cell Microbiol {{DOI|10.1111/cmi.12151}}</ref> The MTRAP monomers interact via their tandem TSR domains with the Sema domains of a Semaphorin-7A homodimer. |
|||
The motile forms have their own stage specific cell surface TRAP family member: TRAP and S6 (also known as TREP) occur on the sporozoites; CTRP is found on the ookinetes; MTRAP is expressed in the merozoites; and TLP is present on both sporozoites and merozoites. Other members of this family are the proteins CSP, SPATR, TRSP, WARP and PTRAMP. Roles for several of these proteins has been discovered: TRAP is critical for sporozoite invasion of the mosquito salivary glands, infection of mammalian liver and sporozoite gliding motility; CTRP is required for invasion of the mosquito midgut; and S6 is important for both sporozoite gliding motility and invasion of mosquito salivary glands. TLP has a role in sporozoite cell traversal. The cytoplasmic tail of TRAP is essential for gliding motility and invasion of the mosquito's salivary glands. Both the TSR and A domains of TRAP are required for the invasion of the mosquito salivary glands. Penetration of the mammalian hepatocytes however requires the TSR, the A domain and the cytoplasmic tail. In contrast only the A domains of CTRP are essential for infectivity by the ookinete. |
|||
PfTRAMP is localised to the base of the rhopteries.<ref name=Pandey2013>Pandey AK, Reddy KS, Sahar T, Gupta S, Singh H, Reddy EJ, Asad M, Siddiqui FA, Gupta P, Singh B, More KR, Mohmmed A, Chitnis CE, Chauhan VS, Gaur D (2013) Identification of a potent combination of key ''Plasmodium falciparum'' merozoite antigens that elicit strain-transcending parasite-neutralizing antibodies. Infect Immun 81(2):441-451 doi: 10.1128/IAI.01107-12</ref> |
|||
The protein PfTCTP causes the release of histamine in the host.<ref name=Pelleau2012>Pelleau S, Diop S, Badiane MD, Vitte J, Beguin P, Nato F, Diop BM, Bongrand P, Parzy D, Jambou R (2012) Enhanced basophil reactivities during severe malaria and their relationship with the plasmodial histamine releasing factor PfTCTP. Infect Immun</ref> This protein also activates the [[basophil]]s. |
|||
The [[cysteine rich protective antigen]] appears to play a role in this process.<ref name=Dreyer2012>Dreyer AM, Matile H, Papastogiannidis P, Kamber J, Favuzza P, Voss TS, Wittlin S, Pluschke G (2012) Passive immunoprotection of ''Plasmodium falciparum''-infected mice designates the CyRPA as candidate malaria vaccine antigen. J Immunol</ref> |
|||
The ribosomal phosphoprotein [[RPLP0|P0]] also seems to be involved in the invasion process.<ref name=Chatterjee2000>Chatterjee S, Singh S, Sohoni R, Kattige V, Deshpande C, Chiplunkar S, Kumar N, Sharma S (2000) Characterization of domains of the phosphoriboprotein P0 of ''Plasmodium falciparum''. Mol Biochem Parasitol 107(2) 143-154</ref> |
|||
A double C2 domain (DOC2) protein appears to be involved in the invasion of the erythrocyte.<ref name=Farrell2012>{{cite journal |author=Farrell A, Thirugnanam S, Lorestani A, Dvorin JD, Eidell KP, Ferguson DJ, Anderson-White BR, Duraisingh MT, Marth GT ''et al.'' |year=2012 |title=A DOC2 protein identified by mutational profiling is essential for apicomplexan parasite exocytosis |journal=Science |volume=335 |issue=6065 |pages=218–221 |doi=10.1126/science.1210829 |pmid=22246776 |pmc=3354045}}</ref> DOC2 proteins recruit the membrane fusion machinery an essential part of the [[Calcium|Ca]]<sup>2+</sup>-dependent exocytosis mechanism.<ref name=Duncan2000>{{cite journal |author=Duncan RR, Shipston MJ, Chow RH |year=2000 |title=Double C2 protein. A review |journal=Biochimie |volume=82 |issue=5 |pages=421–426 |doi=10.1016/S0300-9084(00)00214-5 |pmid=10865129}}</ref> These proteins have a Munc13-interacting domain and tandem C2s (designated C2A and C2B) which are connected by a short polar linker. The C2 domains bind [[phospholipid]]s in a Ca<sup>2+</sup>-dependent manner. Elucidating their precise role in erythrocyte invasion requires further work. |
|||
In ''[[Plasmodium vivax]]'' a number of [[tryptophan]] rich antigens are involved in erythrocyte invasion.<ref name=Tyagi2012>Tyagi RK, Sharma YD (2012) Erythrocyte binding activity displayed by a selective group of ''Plasmodium vivax'' tryptophan rich antigens is inhibited by patients' antibodies" ''PLoS One'' 7(12) e50754. {{DOI|10.1371/journal.pone.0050754}}</ref> Homologs of these proteins are found in ''P. falciparum'' - tryptophan-threonine rich antigen (PfTryThrA) and merozoite associated tryptophan rich antigen (PfMaTrA) and ''[[Plasmodium yoelii]]''. These proteins also seem to be involved in the invasion process. |
|||
In ''P. vivax'' the asparagine rich protein has been cloned.<ref name=Moreno-Pérez2013>Moreno-Pérez DA, Saldarriaga A, Patarroyo MA (2013) Characterizing PvARP, a novel ''Plasmodium vivax'' antigen. Malar J 12:165. {{DOI|10.1186/1475-2875-12-165}}</ref> It is a 281-residue-long molecule, which is encoded by a single exon and has an N-terminal secretion signal in addition to a tandem repeat region. This protein is expressed in mature schizonts and is located on the merozoite surface and appears to accumulate towards the apical pole. |
|||
Another protein that appears to be involved in the invasion process is PfDBLMSP (PF10_0348).<ref name=Wickramarachchi2009>Wickramarachchi T, Cabrera AL, Sinha D, Dhawan S, Chandran T, Devi YS, Kono M, Spielmann T, Gilberger TW, Chauhan VS, Mohmmed A (2009) A novel ''Plasmodium falciparum'' erythrocyte binding protein associated with the merozoite surface, PfDBLMSP. Int J Parasitol 39(7):763-773</ref> This protein has a predicted signal sequence, a central Duffy binding-like (DBL) domain and a secreted polymorphic antigen associated with merozoites (SPAM) domain in its C-terminal half. The transcription and translation of this gene is up regulated specifically in schizonts, similar to other merozoite proteins involved in invasion of erythrocytes. This protein seems to be under selection pressure. |
|||
====Invasion==== |
|||
The process of invasion is partly understood. The merozoite proteins forms a tight junction (the moving junction complex) with some of the erythrocyte membrane proteins.<ref name=Curtidor>Curtidor H, Patiño LC, Arévalo-Pinzón G, Vanegas M, Patarroyo ME, Patarroyo MA (2013) ''Plasmodium falciparum'' rhoptry neck protein 5 peptides bind to human red blood cells and inhibit parasite invasion. Peptides pii: S0196-9781(13)00268-4. {{DOI|10.1016/j.peptides.2013.07.028}}</ref> The attached merozoite proteins are then moved posteriorly by an actin-myosin motor. The net effect of this process is to drive the merozoite into erythrocyte. |
|||
Some details of the invasion process are known.<ref name=Lamarque2011>{{cite journal |author=Lamarque M |year=2011 |title=The RON2-AMA1 interaction is a critical step in moving junction-dependent invasion by apicomplexan parasites |journal=PLoS Pathog |volume=7 |issue=2 |page=e1001276 |author-separator=, |author2=Besteiro S |author3=Papoin J |author4=Roques M |author5=Vulliez-Le Normand B |author6=Morlon-Guyot J |author7=Dubremetz JF |author8=Fauquenoy S |author9=Tomavo S |display-authors=9 |doi=10.1371/journal.ppat.1001276 |editor1-last=Soldati-Favre |editor1-first=Dominique |first10=Bart W. |first11=Clemens H.}}</ref> The rhoptery protein RON2 is inserted into the erythrocyte membrane. The protein AMA1 secreted from microneme then binds to RON2. RON2 forms part of a macromolecular complex which includes RON2, RON 4, RON5 and RON8. The protein PfRON2 via its C-terminal as well as its central cysteine rich domain interacts with PfAMA1.<ref name=Hossain2011>{{cite journal |author=Hossain ME, Dhawan S, Mohmmed A |year=2011 |title=The cysteine-rich regions of ''Plasmodium falciparum'' RON2 bind with host erythrocyte and AMA1 during merozoite invasion |journal=Parasitol Res |pmid=22033736 |doi=10.1007/s00436-011-2690-z |volume=110 |issue=5 |pages=1711–21}}</ref> |
|||
The membrane proximal domain of the AMA1 protein is responsible for direct binding to erythrocytes.<ref name=Tonkin2013>Tonkin ML, Crawford J, Lebrun ML, Boulanger MJ (2013) ''Babesia divergens'' and ''Neospora caninum'' apical membrane antigen 1 structures reveal selectivity and plasticity in apicomplexan parasite host cell invasion" ''Protein Sci'' 22(1) 114-127 {{DOI|10.1002/pro.2193}}</ref> |
|||
The invasion process appears to be ATP dependent<ref name="Levano-Garcia2010">{{cite journal |author=Levano-Garcia J, Dluzewski AR, Markus RP, Garcia CR |year=2010 |title=Purinergic signalling is involved in the malaria parasite ''Plasmodium falciparum'' invasion to red blood cells |journal=Purinergic Signal |volume=6 |issue=4 |pages=365–372 |doi=10.1007/s11302-010-9202-y |pmid=21437007 |pmc=3033500}}</ref> and may involve a purogenic signalling pathway. |
|||
A protein that appears to be unique to the genus ''Plasmodium'' - RON12 has been described.<ref name=Knuepfer2012>Knuepfer E, Suleyman O, Dluzewski AR, Straschil U, O'Keeffe AH, Ogun SA, Green JL, Grainger M, Tewari R, Holder AA (2013) RON12, a novel ''Plasmodium''-specific rhoptry neck protein important for parasite proliferation. Cell Microbiol {{DOI|10.1111/cmi.12181}}</ref> RON12 lacks membrane anchors and is a major soluble component of the nascent parasitophorous vacuole. Most of the secretion of RON12 occurs late during invasion (after parasite internalisation) thus allowing accumulation in the fully formed parasitophorous vacuole. A small proportion of RON12 appears to be present in the moving junction. RON12 does not appear to be essential but its deletion reduces parasite proliferation. |
|||
The motor behind the invasion process is an actinomyosin motor complex that is assembled below the parasite's plasma |
|||
membrane.<ref name=Ridzuan2012>{{cite journal |last1=Ridzuan |first1=MA |last2=Moon |first2=RW |last3=Knuepfer |first3=E |last4=Black |first4=S |last5=Holder |first5=AA |last6=Green |first6=JL |year=2012 |title=Subcellular location, phosphorylation and assembly into the motor complex of GAP45 during ''Plasmodium falciparum'' schizont development |journal=PLoS ONE |volume=7 |issue=3 |page=e33845 |doi=10.1371/journal.pone.0033845 |editor1-last=Langsley |editor1-first=Gordon}}</ref> This complex includes [[myosin]], myosin tail domain interacting protein and glideosome associated proteins 45 and 50. It is anchored on the inner membrane complex which underlies the cell membrane. Myosin, myosin tail domain interacting protein and GAP45 first form a complex that then associates with GAP50. GAP45 is phosphorylated by calcium dependent protein kinase 1 on a number of serine residues. Removal of these residues does not appear to affect the assembly of this complex. This complex may have other function in addition to its role erythrocyte invasion. |
|||
The invasion process requires a coupling of the actin-[[myosin]] motor to the surface receptors. The myosin molecule involved belongs to the single-headed class XIV myosin. For the thromobospondin related anonymous protein on the sporozoites, [[aldolase]] which can bind actin forms this connection.<ref name=Pal-Bhowmick2012>Pal-Bhowmick I, Andersen J, Srinivasan P, Narum DL, Bosch J, Miller LH (2012) Binding of aldolase and glyceraldehyde-3-phosphate dehydrogenase to the cytoplasmic tails of ''Plasmodium falciparum'' merozoite Duffy binding-like and reticulocyte homology ligands. MBio. 2012 Sep 18;3(5). pii: e00292-12. {{DOI|10.1128/mBio.00292-12}}</ref> This connection requires [[tryptophan]] and negatively charged amino acids in the ligand's cytoplasmic tail. PfRH2b also binds aldolase with its cytoplasmic tail. This binning requires an aromatic amino acid ([[phenylalanine]] or [[tyrosine]]) rather than tryptophan again also in the context of negatively charged amino acids. PfLRH2a does not bind aldolase. A second protein [[glyceraldehyde-3-phosphate dehydrogenase]] can also bind actin. It is capable of biding the cytoplasmic tails of some of the PfRh and Duffy binding ligands in an aromatic amino acid dependent manner. |
|||
===Trophozoite=== |
|||
After invading the erythrocyte, the parasite loses its specific invasion organelles ([[apical complex]] and surface coat) and de-differentiates into a round [[trophozoite]] located within a parasitophorous vacuole in the red blood cell cytoplasm. The young [[trophozoite]] (or "ring" stage, because of its morphology on stained blood films) grows substantially before undergoing schizogonic division.<ref name="1.5">{{cite web |
|||
|title=Malaria eModule - ASEXUAL ERYTHROCYTIC STAGES |
|||
|url=http://www.impact-malaria.com/FR/EPS/Formations_et_cours_internationaux/Formation_de_la_Liverpool_School_LSTMH/cours_liverpool/Unit_1/1_5.html}}{{dead link|date=October 2013}}</ref> |
|||
During asexual development the parasite increases in size to ~50% of the uninfected erythrocyte volume: the infected erythrocyte volume remains relatively constant.<ref name=Hanssen2011>{{cite journal |author=Hanssen E, Knoechel C, Dearnley M, Dixon MW, Le Gros M, Larabell C, Tilley L |year=2011 |title=Soft X-ray microscopy analysis of cell volume and hemoglobin content in erythrocytes infected with asexual and sexual stages of ''Plasmodium falciparum'' |journal=J Struct Biol |pmid=21945653 |doi=10.1016/j.jsb.2011.09.003 |volume=177 |issue=2 |pages=224–32 |pmc=3349340}}</ref> Haemoglobin content gradually decreases but its concentration remains constant until the early trophozoite stage when it decreases by 25%. It then remains constant again until just prior to rupture. During early sexual development the gametocyte has a similar morphology to a trophozoite but subsequently undergoes a dramatic shape change. |
|||
The parasite's presence within the erythrocyte induces changes in the properties of the host cell. Relative membrane deformability is less than 10% of uninfected erythrocytes.<ref name=Fedosov2011>{{cite journal |author=Fedosov DA, Lei H, Caswell B, Suresh S, Karniadakis GE |year=2011 |title=Multiscale modeling of red blood cell mechanics and blood flow in malaria |journal=PLoS Comput Biol |volume=7 |issue=12 |page=e1002270 |pmid=22144878 |doi=10.1371/journal.pcbi.1002270 |pmc=3228770 |editor1-last=Beard |editor1-first=Daniel A}}</ref> This change may contribute to the capillary occlusions that occurs in this disease. The deformability of the membrane is also dependent on the temperature and decreases with increased temperature. Deformability is reduced by a factor of 3-4 between 37 and 41 degrees Celsius. The fever that is commonly found in malaria may also contribute via this mechanism to capillary occlusion. The stiffness of the erythrocyte membrane increases as the parasite matures. The overall effect of these chances are to transform the erythrocyte from its normal biconcave shape into a [[parachute]] like structure. This change is apparent at high pressure rather than at low. Transition occurs at flow rates of ~65 µm per [[second]]. The mechanism of these changes are not known but changes in ATP consumption or alterations to the erythrocytes' [[spectrin]] framework may be important. |
|||
Within the red blood cell, the parasite metabolism depends greatly on the digestion of [[hemoglobin]]. A set of enzymes known as [[plasmepsin]]s which are [[protease|aspartic acid proteases]] are used to degrade [[hemoglobin]]. The parasite digests 70-80% of the erythrocyte's haemoglobin<ref name="Francis1997">{{cite journal |coauthors=Sullivan, D. J.; Goldberg, D. E. |year=1997 |title=Hemoglobin metabolism in the malaria parasite ''Plasmodium falciparum'' |journal=Ann. Review Micro. |volume=51 |issue=1 |pages=97–123 |doi=10.1146/annurev.micro.51.1.97 |pmid=9343345 |last1=Francis |first1=SE |last2=Sullivan Dj |first2=Jr |last3=Goldberg |first3=DE}}</ref> but utilizes only ~15% in ''de novo'' protein synthesis.<ref name="Krugliak2002">{{cite journal |last=Krugliak |first=M. |coauthors=Zhang, J.; Ginsburg, H. |year=2002 |title=Intraerythrocytic ''Plasmodium falciparum'' utilizes only a fraction of the amino acids derived from the digestion of host cell cytosol for the biosynthesis of its proteins |journal=Molecular and Biochemical Parasitology |pmid=11814576 |volume=119 |issue=2 |pages=249–256 |doi=10.1016/S0166-6851(01)00427-3}}</ref> Intraerythrocytic ''Plasmodium falciparum'' utilizes only a fraction of the amino acids derived from the digestion of host cell cytosol for the biosynthesis of its proteins. The excess amino acids are exported from the infected erythrocyte by new transport pathways created by the parasite.<ref name="Ginsburg1983">{{cite journal |last=Ginsburg |first=H. |coauthors=Krugliak, M.; Eidelman, O.; Cabantchik, Z.I. |year=1983 |title=New permeability pathways induced in membranes of ''Plasmodium falciparum'' |journal=Mol. Biochem. Parasitol. |volume=8 |issue=2 |pages=177–190 |pmid=6348537 |doi=10.1016/0166-6851(83)90008-7}}</ref> The reason proposed for this apparently excessive digestion of haemoglobin is the colloid-osmotic hypothesis<ref name="Lew2003">{{cite journal |coauthors=Tiffert, T.; Ginsburg, H. |year=2003 |title=Excess hemoglobin digestion and the osmotic stability of ''Plasmodium falciparum''-infected red blood cells |journal=Blood |volume=101 |issue=10 |pages=4189–4194 |doi=10.1182/blood-2002-08-2654 |pmid=12531811 |last1=Lew |first1=VL |last2=Tiffert |first2=T |last3=Ginsburg |first3=H}}</ref> which suggests that the digestion of haemoglobin increases the osmotic pressure within the infected erythrocyte leading to its premature rupture and subsequent death of the parasite. To avoid this fate much of the haemoglobin is digested and exported from the erythrocyte. This hypothesis has been experimentally confirmed.<ref name="Esposito2008">{{cite journal |year=2008 |title=FRET Imaging of Hemoglobin Concentration in Plasmodium falciparum-Infected Red Cells |journal=[[PLoS ONE]] |volume=3 |issue=11 |pages=e3780 |doi=10.1371/journal.pone.0003780 |pmid=19023444 |last1=Esposito |first1=A |last2=Tiffert |first2=T |last3=Mauritz |first3=JM |last4=Schlachter |first4=S |last5=Bannister |first5=LH |last6=Kaminski |first6=CF |last7=Lew |first7=VL |pmc=2582953 |display-authors=1 |editor1-last=Schnur |editor1-first=Joel M.}}</ref> |
|||
Infected erythrocytes are often sequestered in various human tissues or organs, such as the heart, liver and brain. This is caused by parasite derived cell surface proteins being present on the red blood cell membrane and it is these proteins that bind to receptors on human cells. Sequestration in the brain causes cerebral malaria, a very severe form of the disease, which increases the victim's likelihood of death. |
|||
The parasite can also alter the morphology of the red blood cell causing knobs on the erythrocyte membrane. |
|||
Erythrocyte invasion and growth leads to activation of several distinct anion channels and a non-selective Ca2+-permeable cation channel.<ref name=Lang2004>{{cite journal |author=Lang F, Lang PA, Lang KS, Brand V, Tanneur V, Duranton C, Wieder T, Huber SM |year=2004 |title=Channel-induced apoptosis of infected host cells-the case of malaria |journal=Pflugers Arch |volume=448 |issue=3 |pages=319–324 |doi=10.1007/s00424-004-1254-9 |pmid=15042371}}</ref> The non-selective cation channel's activation allows entry of Ca2+ and Na+. Absence of the channels is incompatible with pathogen survival. Although the mechanism of activation of these channels is not know it is presumed to be due to oxidation stree generated by the parasite because similar or identical channels are activated by oxidation of non-infected erythrocytes. Ca2+ entry stimulates an intraerythrocytic [[scramblase]] that facilitates bi-directional [[phospholipid]] migration across the bilayer. This results in an alternation of the cell membrane's [[phosphatidylserine]] asymmetry. Exposure of phosphatidylserine at the outer surface of the cell membrane is followed by binding to phosphatidylserine receptors on [[macrophage]]s and the subsequent [[phagocytosis]] of the affected erythrocyte. It appears that the parasite because of its growth requirements is in a race to complete its life cycle before the infected erythrocyte is phagocytosed. |
|||
===Schizont=== |
|||
At the schizont stage, the parasite replicates its DNA multiple times without cellular segmentation. These schizonts then undergo cellular segmentation and differentiation to form roughly 16-18 merozoite cells in the erythrocyte.<ref name="1.5"/> The rhoptries are formed mainly between second and fourth nuclear divisions; the micronemes between the end of the fourth nuclear division and merozoite separation from the residual body.<ref name=Margos2004>Margos G, Bannister LH, Dluzewski AR, Hopkins J, Williams IT, Mitchell GH (2004) Correlation of structural development and differential expression of invasion-related molecules in schizonts of ''Plasmodium falciparum''" ''Parasitology'' 129(3) 273-287</ref> The dense granules are formed mainly after the micronemes. |
|||
The merozoites burst from the red blood cell, and proceed to infect other erythrocytes. The parasite is in the bloodstream for roughly 60 seconds before it has entered another erythrocyte.<ref name="cowman">{{cite journal |
|||
|last=Cowman |
|||
|title=Invasion of Red Blood Cells by Malaria Parasites |
|||
|journal=Cell |
|||
|volume=124 |
|||
|issue=4 |
|||
|pages=755–766 |
|||
|date=24 February 2006 |
|||
|doi=10.1016/j.cell.2006.02.006 |
|||
|pmid=16497586 |
|||
|first1=AF |
|||
|last2=Crabb |
|||
|first2=BS}}</ref> |
|||
This infection cycle occurs in a highly synchronous fashion, with roughly all of the parasites throughout the blood in the same stage of development. This precise clocking mechanism has been shown to be dependent on the human host's own [[circadian rhythm]].<ref name="1.5.2">{{cite web |
|||
|title=Malaria eModule - SYNCHRONICITY |
|||
|url=http://www.impact-malaria.com/FR/EPS/Formations_et_cours_internationaux/Formation_de_la_Liverpool_School_LSTMH/cours_liverpool/Unit_1/1_5_2.html}}{{dead link|date=October 2013}}</ref> Specifically, human body temperature changes as a result of the circadian rhythm, seem to play a role in the development of ''[[Plasmodium falciparum|P. falciparum]]'' within the erythrocytic stage. |
|||
The synchronicity of the erythrocytic cycle is at least in part dependent on [[melatonin]] secretion by the host. A mechanism for this has been proposed.<ref name="Alves2010">{{cite journal |author=Alves E, Bartlett PJ, Garcia CR, Thomas AP |year=2010 |title=Melatonin and IP3-induced Ca2+ release from intracellular stores in the malaria parasite Plasmodium falciparum within infected red blood cells. J. Biol |journal=Chem.}}</ref> Melatonin can activate phospholipase C which acts to generate [[inositol trisphosphate]] (IP3) which opens IP3 sensitive [[calcium]] channels in the [[endoplasmic reticulum]]. The released calcium in its turn controls the cycle through mechanisms that have yet to be understood. |
|||
===Gametocyte differentiation=== |
===Gametocyte differentiation=== |
||
During the erythrocytic stage, some merozoites develop into male and female gametocytes |
During the erythrocytic stage, some merozoites develop into male and female gametocytes - a process is called gametocytogenesis.<ref name="1.6.1">{{cite web |
||
|title=Malaria eModule - GAMETOCYTOGENESIS |
|||
| last = |
|||
|url=http://www.impact-malaria.com/FR/EPS/Formations_et_cours_internationaux/Formation_de_la_Liverpool_School_LSTMH/cours_liverpool/Unit_1/1_6_1.html}}{{dead link|date=October 2013}}</ref> The specific factors and causes underlying this sexual differentiation are largely unknown. The base rate appears to be ~10% in laboratory strains but this is subject to many influences. One of the earliest suggestions in this area was made by Sinton who proposed that 'stress' might influence the rate of gametogensis.<ref name=Sinton1938>Sinton JA (1938) The action of Atebrin upon gametocytes of ''Plasmodium falciparum''. Riv Malariol 17:305-330</ref> There is some evidence from other workers to support this hypothesis.<ref name=Reece2008>Reece SE, Drew DR, Gardner A (2008) Sex ratio adjustment and kin discrimination in malaria parasites" ''Nature'' 453(7195) 609-614</ref> Factors that have been reported to influence this rate include host erythrocyte age, hypoxia and exposure to schizonticidal drugs. |
|||
| first = |
|||
| authorlink = |
|||
| coauthors = |
|||
| title = Malaria eModule - GAMETOCYTOGENESIS |
|||
| work = |
|||
| publisher = |
|||
| date = |
|||
| url = http://www.impact-malaria.com/FR/EPS/Formations_et_cours_internationaux/Formation_de_la_Liverpool_School_LSTMH/cours_liverpool/Unit_1/1_6_1.html |
|||
| format = |
|||
| doi = |
|||
| accessdate = }}</ref> The specific factors and causes underlying this sexual differentiation are largely unknown. These gametocytes take roughly 8-10 days to reach full maturity. Note that the gametocytes remain within the erythrocytes until taken up by the [[mosquito]] host. |
|||
Gametocytes take roughly 8–10 days to reach full maturity and are metabolically active. The gametocytes remain within the erythrocytes until taken up by the [[mosquito]] host. The osmiophilic bodies, present in both male and female gametocytes but more abundant in the latter, are involved in the parasites' escape from the red blood cell during gametogenesis.<ref name=Ponzi2009>Ponzi M, Siden-Kiamos I, Bertuccinin L, Curra C, Kroeze H, Camarda G, Pace T, Franke-Fayard B, Laurentino EC, Louis K, Waters AP, Janse CJ, Alano P (2009) Egress of ''Plasmodium berghei'' gametes from their host erythrocyte is mediated by MDV-1/PEG3 protein. Cell Microbiol 11:1272-1288</ref><ref>Lal K, Delves MJ, Bromley E, Wastling JM, Tomley FM, Sinden RE (2009) ''Plasmodium'' male development gene-1 (mdv-1) is important for female sexual development and identifies a polarised plasma membrane during zygote development. Int J Parasitol 39:755-761</ref> |
|||
===Mosquito stage=== |
|||
''[[Plasmodium falciparum|P. falciparum]]'' is taken up by the female ''[[Anopheles]]'' [[mosquito]] as it takes its bloodmeal from an infected human. |
|||
There is considerable variation in the ratio of female to male gametocytes between strains.<ref name=Robert1996>{{cite journal |last1=Robert |first1=V |last2=Read |first2=AF |last3=Essong |first3=J |last4=Tchuinkam |first4=T |last5=Mulder |first5=B |last6=Verhave |first6=JP |last7=Carnevale |first7=P |year=1996 |title=Effect of gametocyte sex ratio on infectivity of ''Plasmodium falciparum'' to ''Anopheles gambiae'' |journal=Trans R Soc Trop Med Hyg |volume=90 |issue=6 |pages=621–624 |doi=10.1016/S0035-9203(96)90408-3 |pmid=9015496}}</ref> Typically there are four female gametocytes to each male gametocyte. Mathematical modelling suggests that this may be due to differences in male fecundity.<ref name=Teboh-Ewungkem2012>Teboh-Ewungkem MI, Wang M (2012) Male fecundity and optimal gametocyte sex ratios for ''Plasmodium falciparum'' during incomplete fertilization. J Theor Biol</ref> |
|||
====''Gametogenesis''==== |
|||
The protein Pfg377 has been shown to be localised to the osmophilic bodies of the gametocytes.<ref name=Sannella2012>Sannella AR, Olivieri A, Bertuccini L, Ferre F, Severini C, Pace T, Alano P (2012) Specific tagging of the egress-related osmiophilic bodies in the gametocytes of ''Plasmodium falciparum''. Malar J 11(1) 88</ref> |
|||
Upon being taken up by the [[mosquito]], the gametocytes leave the erythrocyte shell and differentiate into gametes. The female gamete maturation process entails slight morphological changes, as it becomes enlarged and spherical. On the other hand, the male gamete maturation involves significant morphological development. The male gamete's [[DNA]] divides three times to form eight nuclei. Concurrently, eight [[flagella]] are formed. Each [[flagella]] pairs with a nucleus to form a microgamete, which separates from the parasite cell. This process is referred to as [[exflagellation]]. |
|||
The development of gametocytes is associated with down regulation of the erythrocyte membrane protein 1 (the product of the ''var'' genes) and loss of cytoadhesion.<ref name="Tibúrcio2012">Tibúrcio M, Silvestrini F, Bertuccini L, Sander A, Turner L, Lavstsen T, Alano P (2012) Early gametocytes of the malaria parasite Plasmodium falciparum specifically remodel the adhesive properties of infected erythrocyte surface. Cell Microbiol {{DOI|10.1111/cmi.12062}}</ref> |
|||
Gametogenesis has been shown to be caused by: 1) a sudden drop in temperature upon leaving the human host, 2) a rise in pH within the [[mosquito]], and 3) [[xanthurenic acid]] within the mosquito.<ref name="billker">{{cite journal |
|||
| last = Billker |
|||
| first = |
|||
| authorlink = |
|||
| coauthors = |
|||
| title = Identification of xanthurenic acid as the putative inducer of malaria development in the mosquito |
|||
| journal = Nature |
|||
| volume = 392 |
|||
| issue = |
|||
| pages = 289–292 |
|||
| publisher = |
|||
| location = |
|||
| date = March 19, 1998 |
|||
| doi =10.1038/32667 |
|||
| id = |
|||
| accessdate = }}</ref> |
|||
The CPW-WPC protein family, named after the unique WxC repeat domains, is highly conserved among Plasmodium species. It is transcribed in gametocytes and CPW-WPC proteins are expressed in zygote/ookinete-stage parasites.<ref name=Kangwanrangsan2013>Kangwanrangsan N, Tachibana M, Jenwithisuk R, Tsuboi T, Riengrojpitak S, Torii M, Ishino T (2013) A member of the CPW-WPC protein family is expressed in and localized to the surface of developing ookinetes. Malar J 12(1) 129</ref> These do not appear to be essential genes. |
|||
====''Fertilization''==== |
|||
====Sexual differentiation==== |
|||
Fertilization of the female gamete by the male gamete occurs rapidly after gametogenesis. The fertilization event produces a [[zygote]]. The [[zygote]] then develops into an [[ookinete]]. The [[zygote]] and [[ookinete]] are the only [[diploid]] stages of ''[[Plasmodium falciparum|P. falciparum]]''. |
|||
Sexual parasite development is controlled by a [[DEAD box]] [[RNA]] [[helicase]] of the [[DDX6 family]], termed DOZI.<ref name="Müller2011"/> |
|||
====''Ookinete''==== |
|||
The Puf2 gene, a member of the [[Puf]] family of transcriptional regulators, has been shown to be involved in gamete formation.<ref name="Miao2010 ">{{cite journal |author=Miao J, Li J, Fan Q, Li X, Li X, Cui L |year=2010 |title=The Puf-family RNA-binding protein PfPuf2 regulates sexual development and sex differentiation in the malaria parasite ''Plasmodium falciparum'' |journal=J. Cell Sci. |doi=10.1242/jcs.059824 |volume=123 |issue=7 |pages=1039}}</ref> |
|||
The diploid [[ookinete]] is an invasive form of ''[[Plasmodium falciparum|P. falciparum]]'' within the [[mosquito]]. It traverses the [[peritrophic membrane]] of the [[mosquito]] midgut and cross the midgut epithelium. Once through the epithelium, the [[ookinete]] enters the basil lamina, and forms an [[oocyst]]. |
|||
[[FACT (biology)|FACT]] (facilitates chromatin transcription) is a dimeric complex of two [[protein]]s - [[SUPT16H|SPT16]] and [[SSRP1]] - which acts as a [[histone]] [[chaperone (protein)|chaperone]] in the (dis)assembly of [[nucleosome]] (and [[chromatin]]) structure during [[transcription (genetics)|transcription]] and DNA replication.<ref name=Laurentino2011>Laurentino EC, Taylor S, Mair GR, Lasonder E, Bartfai R, Stunnenberg HG, Kroeze H, Ramesar J, Franke-Fayard B, Khan SM, Janse CJ, Waters AP (2011) Experimentally controlled down regulation of the histone chaperone FACT in ''Plasmodium berghei'' reveals that it is critical to male gamete fertility. Cell Microbiol {{doi|10.1111/j.1462-5822.2011.01683.x}}</ref> It is an essential gene in ''Plasmodium''. Changing its promoter to one expressed only in the blood stages leads to changes in the male gametocytes. The mutant gametocytes have delayed DNA replication and gametocyte formation. Male gamete fertility is strongly reduced. Female gametocytes appear to be normal. When successful fertilization is achieved, the ookinetes generate oocysts that arrest early in development and fail to enter sporogony. |
|||
During the [[ookinete]] stage, genetic recombination can occur. This takes place if the [[ookinete]] was formed from male and female gametes derived from different populations. This can occur if the human host contained multiple populations of the parasite, or if the [[mosquito]] fed from multiple infected individuals within a short time-frame. |
|||
The proteins [[cell division cycle protein 20]] and its homologue, CDC20 homologue 1 are central to the cell cycle activating the [[anaphase-promoting complex]]/[[cyclosome]] (APC/C) in mitosis and facilitating degradation of mitotic APC/C substrates.<ref name=Guttery2012>{{cite journal |author=Guttery DS, Ferguson DJ, Poulin B, Xu Z, Straschil U, Klop O, Solyakov L, Sandrini SM, Brady D ''et al.'' |year=2012 |title=A Putative Homologue of CDC20/CDH1 in the malaria parasite is essential for male gamete development |journal=PLoS Pathog |volume=8 |issue=2 |page=e1002554 |doi=10.1371/journal.ppat.1002554 |editor1-last=Soldati-Favre |editor1-first=Dominique}}</ref> A single homolog of this gene has been identified in ''[[Plasmodium berghei]]''. It appears to be essential in male gametogensis but not for asexual reproduction. Blockage occurs at the nuclear spindle/kinetochore stage. |
|||
====''Sporogony''==== |
|||
A gametocyte development 1 gene (Gdv1) which encodes a perinuclear protein has been identified.<ref>Eksi S, Morahan BJ, Haile Y, Furuya T, Jiang H, Ali O, Xu H, Kiattibutr K, Suri A, Czesny B, Adeyemo A, Myers TG, Sattabongkot J, Su XZ, Williamson KC (2012) ''Plasmodium falciparum'' Gametocyte development 1 (Pfgdv1) and gametocytogenesis early gene identification and commitment to sexual development" ''PLoS Pathog'' 8(10) e1002964. {{DOI|10.1371/journal.ppat.1002964}}</ref> Its mechanism of action is not known. Homologues of this gene have been found in ''[[Plasmodium vivax]]'', ''[[Plasmodium knowlesi]]'' and ''[[Plasmodium gallinaceum]]''. |
|||
Over the period of a 1-3 weeks, the [[oocyst]] grows to a size of tens to hundreds of micrometres. During this time, multiple nuclear divisions occur. After [[oocyst]] maturation is complete, the [[oocyst]] divides to form multiple haploid sporozoites. Immature sporozoites break through the [[oocyst]] wall into the haemolymph. The sporozoites then migrate to the salivary glands an complete their differentiation. Once mature, the sporozoites can proceed to infect a human host during a subsequent [[mosquito]] bite. |
|||
===Egress from the erythrocyte=== |
|||
This is an essential step in the life cycle. The calcium dependent protein kinase [[PfCDPK5]] which is expressed in the merozoite is essential for this process.<ref>{{cite journal |last1=Dvorin |first1=JD |last2=Martyn |first2=DC |last3=Patel |first3=SD |last4=Grimley |first4=JS |last5=Collins |first5=CR |last6=Hopp |first6=CS |last7=Bright |first7=AT |last8=Westenberger |first8=S |last9=Winzeler |first9=E ''et al.'' |year=2010 |title=A Plant-Like Kinase in Plasmodium falciparum Regulates Parasite Egress From Erythrocytes |journal=Science |volume=328 |issue=5980 |pages=910–912 |display-authors=9 |doi=10.1126/science.1188191 |pmid=20466936 |pmc=3109083 |last10=Blackman |first10=M. J. |last11=Baker |first11=D. A. |last12=Wandless |first12=T. J. |last13=Duraisingh |first13=M. T.}}</ref> Deletion mutations of this gene result in cell arrest in the late schizont stages. Merozoites released from these schizonts are capable of invasion. |
|||
The mechanics of release from the host membrane are partly known.<ref name=Callan-Jones2012>Callan-Jones A, Albarran Arriagada OE, Massiera G, Lorman V, Abkarian M (2012) Red blood Cell membrane dynamics during malaria parasite egress. Biophys J. 2012 Dec 19;103(12) 2475-83. {{DOI|10.1016/j.bpj.2012.11.008}}</ref> Before release a pore opens in the membrane. A distortion of the usual arrengement of the lipids and proteins of the host cell membrane occurs around the pore. This area of altered protein and lipid eventually ruptures. |
|||
Large holes appear in the cytoskeleton ~35 hours post invasion.<ref name=Millholland2011>{{cite journal |author=Millholland MG, Chandramohanadas R, Pizarro A, Wehr A, Shi H, Darling C, Lim CT, Greenbaum DC |year=2011 |title=The malaria parasite progressively dismantles the host erythrocyte cytoskeleton for efficient egress |journal=Mol Cell Proteomics}}</ref> This occurs at the same time as the loss of cytoskeletal adaptor proteins that are part of the junctional complex, including α/β-[[adducin]] and [[tropomyosin]]. This is followed by the proteolysis of many cytoskeletal proteins during egress at ~48 hours post infection. This later proteolysis is mediated by the erythrocyte's own [[calpain]]-1. |
|||
Infected erythrocytes release microvesicles from their surface. They contain a number of parasite antigens associated with host cell membranes and proteins involved in parasite invasion which have potent immunomodulatory properties affecting macrophages and neutrophils.<ref name=Mantel2013>Mantel PY, Hoang AN, Goldowitz I, Potashnikova D, Hamza B, Vorobjev I, Ghiran I, Toner M, Irimia D, Ivanov AR, Barteneva N, Marti M (2013) Malaria-infected erythrocyte-derived microvesicles mediate cellular communication within the parasite population and with the host immune system. Cell Host Microbe 13(5) 521-534 {{DOI|10.1016/j.chom.2013.04.009}}</ref> Their uptake by infected eythrocytes stimulates the production of transmission stage parasites in a dose dependent manner. Their release increases during the asexual parasite cycle particularly prior to parasite egress. A protein - PTP2 - appears to be essential in this process but its role is not yet clear.<ref name=Regev-Rudzki2013>Regev-Rudzki N, Wilson DW, Carvalho TG, Sisquella X, Coleman BM, Rug M, Bursac D, Angrisano F, Gee M, Hill AF, Baum J, Cowman AF (2013) Cell-cell communication between malaria-infected red blood cells via exosome-like vesicles. Cell pii: S0092-8674(13)00504-7. {{DOI|10.1016/j.cell.2013.04.029}}</ref> |
|||
A Gα(q) coupled signaling pathway that results in [[protein kinase C]] mediated loss of the host cytoskeletal protein [[adducin]] and weakening of the cellular cytoskeleton has also been implicated in the egress mechanism.<ref>Millholland MG, Mishra S, Dupont CD, Love MS, Patel B, Shilling D, Kazanietz MG, Foskett JK, Hunter CA, Sinnis P, Greenbaum DC (2013) A host GPCR signaling network required for the cytolysis of infected cells facilitates release of apicomplexan parasites. Cell Host Microbe 13(1) 15-28. {{DOI|10.1016/j.chom.2012.12.001}}</ref> This weaking of the cytoskeletal induces catastrophic Ca<sup>2+</sup> influx mediated by the mechanosensitive cation channel [[TRPC6]]. This in turn which activates host cell's calpain which proteolyzes the host cytoskeleton allowing parasite release. |
|||
Along with the release from the erythrocyte of the merozoites, the now functionless digestive vacuole is also released. These can active complement and are rapidly taken up by the [[Granulocyte|polymorph]]s.<ref name=Dasari2011>{{cite journal |author=Dasari P, Reiss K, Lingelbach K, Baumeister S, Lucius R, Udomsangpetch R, Bhakdi SC, Bhakdi S |year=2011 |title=Digestive vacuoles of ''Plasmodium falciparum'' are selectively phagocytosed by and impair killing function of polymorphonuclear leukocytes |journal=Blood |doi=10.1182/blood-2011-05-353920 |volume=118 |issue=18 |pages=4946–56 |pmid=21911835}}</ref> On ingestion the digestive vacuoles induce a vigorous respiratory burst which drives the cells into a state of functional exhaustion, blunting production of reactive oxygen species and microbicidal activity upon challenge with bacterial pathogens. |
|||
The serine repeat antigen (SERA) multigene family encode a series of proteins with a putative [[papain]]-like [[cysteine]] [[protease]] motif. One of these SERA5 (120 kilo[[Dalton (unit)|Dalton]]s) is produced at the late trophozoite/schizont stage. It is secreted together with other SERAs into the parasitophorous vacuole in an infected erythrocyte where it is cleaved into three fragments: an N-terminal domain (47 kDa), a central domain containing putative papain-like cysteine protease motifs (56 kDa) and a C-terminal domain (18 kDa). This N-terminal fragment is then cleaved in turn into two 25 kDa fragments. These fragments become covalently linked to the C-terminal 18 kDa fragment via disulfide bonding and attach to the merozoite surface. The central fragment is further cleaved to 50 kDa and 6 kDa fragments before being shed to the medium. These proteolytic cleavages are carried out by a [[subtilisin]]-like [[serine]] protease called PfSUB1 and the inhibition of this processing, likewise, results in blockade of merozoite release.<ref name=Arisue2011>{{cite journal |last1=Arisue |first1=N |last2=Kawai |first2=S |last3=Hirai |first3=M |last4=Palacpac |first4=NM |last5=Jia |first5=M |last6=Kaneko |first6=A |last7=Tanabe |first7=K |last8=Horii |first8=T |year=2011 |title=Clues to evolution of the SERA multigene family in 18 ''Plasmodium'' species |journal=PLoS ONE |volume=6 |issue=3 |page=e17775 |doi=10.1371/journal.pone.0017775 |editor1-last=Langsley |editor1-first=Gordon}}</ref> SERA6 may also be involved in schizont rupture and merozoite release from the erythrocyte. Both SERA5 and SERA6 are essential for blood stage parasite viability.<ref name=Ruecker2012>Ruecker A, Shea M, Hackett F, Suarez C, Hirst EM, Milutinovic K, Withers-Martinez C, Blackman MJ (2012) Proteolytic activation of the essential parasitophorous vacuole cysteine protease SERA6 accompanies malaria parasite egress from its host erythrocyte. J Biol Chem</ref> SERA6 is found in parasitophorous vacuole where it is activated by cleavage by the serine protease PfSUB1 just prior to egress. The release of PfSUB1 may be controlled by a calcium flux within the exomemes - storage vesicles within the parasite - of the merozoites.<ref name=Agarwal2012>Agarwal S, Singh MK, Garg S, Chitnis CE, Singh S (2012) Ca(2+) Mediated exocytosis of subtilisin-like protease 1: A key step in egress of ''P. falciparum'' merozoites. Cell Microbiol {{DOI|10.1111/cmi.12086}}</ref> The release may be under the control of a phospholipase C. |
|||
PfSUB1 - which is encoded by the gene PF3D7_0507500 - is released from the exomemes into the parasitophorous vacuole before the merozoite egresses from the erythrocyte. Its release is inhibited by a cyclic GMP dependent protein kinase (encoded by the gene PF3D7_1436600).<ref name=Collins2013>Collins CR, Hackett F, Strath M, Penzo M, Withers-Martinez C, Baker DA, Blackman MJ (2013) Malaria parasite cGMP-dependent protein kinase regulates blood stage merozoite secretory organelle discharge and egress" ''PLoS Pathog'' 9(5) e1003344</ref> Substrates of PfSUB1 include MSP1, MSP6, MSP7, MSRP2, SERA5 and SERA6 and the rhoptry protein RAP1. PfSUB1 acts on MSP1 at three sites to produce 4 smaller proteins. It acts on SERA5 - located within the parasitophorous vacuole - a at one site to produce two smaller proteins. One of these is subsequently cleaved by a second protease. Inhibition of the cyclic GMP dependent protein kinase also inhibits microneme discharge. |
|||
A protein - gamete egress and sporozoite traversal - has been identified that appears to be involved in the egress of male and female gametes from the erythrocyte.<ref name=Talman2011>Talman AM, Lacroix C, Marques SR, Blagborough AM, Carzaniga R, Ménard R, Sinden RE (2011) PbGEST mediates malaria transmission to both mosquito and vertebrate host. Mol Microbiol {{doi|10.1111/j.1365-2958.2011.07823.x}}</ref> It is also involved in sporozoite migration. |
|||
A perforin-like protein (PPLP2) is involved in egress of male gametocytes from the erythrocytes.<ref name=Deligianni2013>Deligianni E, Morgan RN, Bertuccini L, Wirth CC, Silmon de Monerri NC, Spanos L, Blackman MJ, Louis C, Pradel G, Siden-Kiamos I (2013) A perforin-like protein mediates disruption of the erythrocyte membrane during egress of ''Plasmodium berghei'' male gametocytes. Cell Microbiol {{DOI|10.1111/cmi.12131}}</ref> This protein does not appear to be involved in the rupture of the parasitophorous vacuole. Instead of parasites with mutations in this gene produce gametocytes with only one, shared thicker flagellum rather than usual pattern of each male gametocyte forming 8 flagellated gametes. |
|||
Perforin-like protein 1 and perforin-like protein 2 are both transcribed in the blood stage.<ref name=Garg2013>Garg S, Agarwal S, Kumar S, Shams Yazdani S, Chitnis CE, Singh S (2013) Calcium-dependent permeabilization of erythrocytes by a perforin-like protein during egress of malaria parasites. ''[[Nat. Commun.]]'' 4:1736</ref> Perforin-like protein 1 localizes to the red blood cell membrane and parasitophorous vacuolar membrane in mature schizonts following its Ca<sup>2+</sup>-dependent discharge from micronemes. Perforin-like protein 1 has Ca<sup>2+</sup>-dependent permeabilization and membranolytic activities. |
|||
The internal calcium levels rise for two hours pre egress.<ref name=Glushakova2013>Glushakova S, Lizunov V, Blank PS, Melikov K, Humphrey G, Zimmerberg J (2013) Cytoplasmic free Ca2+ is essential for multiple steps in malaria parasite egress from infected erythrocytes. Malar J 12(1) 41</ref> This rise is dependent on internal stores of calcium probably from the endoplasmic reticulum. Inhibition of this rise prevents parasitophorous vacuole swelling and erythrocyte membrane poration. |
|||
Phosphorylation of the reticulocyte homologue protein 2b on residue on [[serine]] 3233 occurs prior to egress.<ref name=Engelberg2013>Engelberg K, Paul AS, Prinz B, Kono M, Ching W, Heincke D, Dobner T, Spielmann T, Duraisingh M, Gilberger TW (2013) Specific phosphorylation of the PfRh2b invasion ligand of ''Plasmodium falciparum''. Biochem J</ref> This is the sole phosphorylation site on this protein. This action is carried put by [[casein kinase 2]] and is cyclic [[Adenosine monophosphate|AMP]] independent, utilizes [[Adenosine triphosphate|ATP]] as well as [[Guanosine triphosphate|GTP]] as [[phosphate]] donors and is inhibited by [[heparin]] and [[tetrabromocinnamic acid]]. This phosphorlation most likely occus most likely occurs before the protein is translocated from the rhoptry neck to the plasma membrane. |
|||
==Mosquito stage== |
|||
''[[Plasmodium falciparum|P. falciparum]]'' is taken up by the female ''[[Anopheles]]'' [[mosquito]] as it takes its bloodmeal from an infected human. Within the midgut human complement remains active for ~1 hour. To protect itself the parasite binds host complement regulator [[factor H]].<ref>Simon N, Lasonder E, Scheuermayer M, Kuehn A, Tews S, Fischer R, Zipfel PF, Skerka C, Pradel G (2012) Malaria parasites co-opt human Factor H to prevent complement-mediated lysis in the mosquito midgut. Cell Host Microbe 13(1) 29-41 {{DOI|10.1016/j.chom.2012.11.013}}</ref> Specifically the gamete surface protein PfGAP50 binds to Factor H and uses the surface bound Factor H to inactivate the complement protein C3b. |
|||
How the parasite evades the mosquito's immune system is not understood. The protein Pfs47 appears to be involved in this process.<ref name=Molina-Cruz2013>Molina-Cruz A, Garver LS, Alabaster A, Bangiolo L, Haile A, Winikor J, Ortega C, van Schaijk BC, Sauerwein RW, Taylor-Salmon E, Barillas-Mury C (2013) The human malaria parasite Pfs47 gene mediates evasion of the mosquito immune system. ''Science''</ref> |
|||
===Gametogenesis=== |
|||
Upon being taken up by the [[mosquito]], the gametocytes leave the erythrocyte shell and differentiate into gametes. The female gamete maturation process entails slight morphological changes, as it becomes enlarged and spherical. On the other hand, the male gamete maturation involves significant morphological development. The male gamete's [[DNA]] divides three times to form eight nuclei. Concurrently, eight [[flagella]] are formed. Each [[flagellum]] pairs with a nucleus to form a microgamete, which separates from the parasite cell - a process known as [[exflagellation]]. |
|||
Gametogenesis has been shown to be caused by:<ref name="billker">{{cite journal |
|||
|last=Billker |
|||
|title=Identification of xanthurenic acid as the putative inducer of malaria development in the mosquito |
|||
|journal=Nature |
|||
|volume=392 |
|||
|issue=6673 |
|||
|pages=289–292 |
|||
|date=March 19, 1998 |
|||
|doi=10.1038/32667 |
|||
|pmid=9521324 |
|||
|first1=O |
|||
|last2=Lindo |
|||
|first2=V |
|||
|last3=Panico |
|||
|first3=M |
|||
|last4=Etienne |
|||
|first4=AE |
|||
|last5=Paxton |
|||
|first5=T |
|||
|last6=Dell |
|||
|first6=A |
|||
|last7=Rogers |
|||
|first7=M |
|||
|last8=Sinden |
|||
|first8=RE |
|||
|last9=Morris |
|||
|first9=HR |displayauthors=9}}</ref> |
|||
* a sudden drop in temperature upon leaving the human host |
|||
* a rise in pH within the [[mosquito]] |
|||
* [[xanthurenic acid]] within the mosquito |
|||
An ATP-binding cassette (ABC) transporter encoded by the gene Pf14_0244 (PfABCG2) on chromosome 14 appears to have some role in the asexual stages, gametocyte stages and in the oocyst.<ref name=Eastman2012>Eastman RT, Pattaradilokrat S, Raj DK, Dixit S, Deng B, Miura K, Yuan J, Tanaka TQ, Johnson RL, Jiang H, Huang R, Williamson K, Lambert LE, Long C, Austin CP, Wu Y, Su XZ (2012) A class of tricyclic compounds blocking malaria oocyst development and transmission. Antimicrob Agents Chemother</ref> |
|||
The process of the formation of flagella is unusual in that they are formed with in the cytoplasm before export.<ref name=Portman2013>Portman N, Foster C, Walker G, Slapeta J (2013) Evidence for intraflagellar transport and apical complex formation in a free living relative of the Apicomplexa. Eukaryot Cell</ref> This pattern is also found in ''[[Chromera velia]]''. |
|||
The movement of the flagellae has been studied.<ref name=>Wilson LG, Carter LM, Reece SE (2013) |
|||
High-speed holographic microscopy of malaria parasites reveals ambidextrous flagellar waveforms. Proc Natl Acad Sci USA 2013 </ref> The flagellae alternate their direction of beating on each stroke: a left to right beat is followed by a right to left beat. |
|||
===Fertilization=== |
|||
Fertilization of the female gamete by the male gamete occurs rapidly after gametogenesis. The motility of the male gametocytes is powered by [[glycolysis]].<ref name=Slavic2011>Slavic K, Delves MJ, Prudencio M, Talman AM, Straschil U, Derbyshire ET, Xu Z, Sinden RE, Mota MM, Morin C, Tewari R, Krishna S, Staines HM (2011) Use of a selective inhibitor to define the chemotherapeutic potential of the plasmodial hexose transporter in different stages of the parasite's life cycle" ''Antimicrob Agents Chemother'' 55:2824-2830</ref> The fertilization event produces a [[zygote]]. The [[zygote]] then develops into an [[ookinete]]. Of all the macrogametes present in the gut only ~2% develop into ookinetes.<ref name=Vaughan1992>Vaughan JA, Noden BH, Beier JC (1992) Population dynamics of ''Plasmodium falciparum'' sporogony in laboratory-infected ''Anopheles gambiae''. J Parasitol 78(4) 716-724</ref> The [[zygote]] and [[ookinete]] are the only [[diploid]] stages of ''[[Plasmodium falciparum|P. falciparum]]''.{{citation needed|date=October 2012}} |
|||
A gene - PBANKA_113980 - essential for the production of viable progeny after meiosis has been identified.<ref name=Ning2013>Ning J, Otto TD, Pfander C, Schwach F, Brochet M, Bushell E, Goulding D, Sanders M, Lefebvre PA, Pei J, Grishin NV, Vanderlaan G, Billker O, Snell WJ (2013) Comparative genomics in ''Chlamydomonas'' and ''Plasmodium'' identifies an ancient nuclear envelope protein family essential for sexual reproduction in protists, fungi, plants, and vertebrates" ''Genes Dev'' 27(10) 1198-1215</ref> This gene is the [[ortholog]] of the gene GEX1 in the plant ''[[Arabidopsis thaliana]]'', the gene Cre06.g280600 in ''[[Chlamydomonas]]'' and the gene KAR5 in the yeast ''[[Saccharomyces cerevisiae]]''. It appears that these genes are involved in nuclear fusion during fertilization. |
|||
===Ookinete=== |
|||
The diploid [[ookinete]] is an invasive form of ''[[Plasmodium falciparum|P. falciparum]]'' within the [[mosquito]]. It traverses the [[peritrophic membrane]] of the [[mosquito]] midgut and cross the midgut epithelium. Once through the epithelium, the [[ookinete]] enters the basal lamina, and forms an [[oocyst]]. Unlike the other invasive stages the ookinete lacks rhopteries. Only 1-2% of ookinites develop into oocyts.<ref name="Vaughan1992"/> |
|||
The processes of maturation and invasion of the mosquito gut are under investigation. Both [[chitinase]] and [[plasmepsin]] 4 (an aspartic acid [[protease]]) are known to be involved in the invasion process.<ref name="Li2010">{{cite journal |author=Li F, Patra KP, Yowell CA, Dame JB, Chin K, Vinetz JM |year=2010 |title=Apical surface expression of aspartic protease plasmepsin 4, a potential transmission-blocking target of the ''Plasmodium'' ookinete |journal=J. Biol. Chem. |pmid=20056606 |doi=10.1074/jbc.M109.063388 |pmc=2832958 |volume=285 |issue=11 |pages=8076–83}}</ref> |
|||
During the [[ookinete]] stage, genetic recombination can occur. This takes place if the [[ookinete]] was formed from male and female gametes derived from different populations. This can occur if the human host contained multiple populations of the parasite, or if the [[mosquito]] fed from multiple infected individuals within a short time-frame. |
|||
A G-actin binding protein has been implicated in this process.<ref name="Hliscs2010">{{cite journal |author=Hliscs M, Sattler J, Tempel W, Artz JD, Dong A, Hui R, Matuschewski K, Schuler H |year=2010 |title=Structure and function of a G-actin sequestering protein with a vital role in malaria oocyst development inside the mosquito vector |journal=J Biol Chem. |pmid=20083609 |doi=10.1074/jbc.M109.054916 |pmc=2857035 |volume=285 |issue=15 |pages=11572–83}}</ref> These proteins - probably acting as dimers - bind actin monomers just before sporogony. |
|||
[[Azithromycin]] has been shown to suppress apicoplast replication at the period of sporozoite production in oocysts.<ref name="Shimizu2010">{{cite journal |last1=Shimizu |first1=S |last2=Osada |first2=Y |last3=Kanazawa |first3=T |last4=Tanaka |first4=Y |last5=Arai |first5=M |author-separator=, |year=2010 |title=Suppressive effect of azithromycin on ''Plasmodium berghei'' mosquito stage development and apicoplast replication |journal=Malar J |volume=9 |issue=1 |page=73 |doi=10.1186/1475-2875-9-73}}</ref> |
|||
The proteins [[enolase]] and [[actin]] are present on the surface of ookinetes but their function there, if any, is unknown.<ref name="Hernández-Romano2011">{{cite journal |author=Hernández-Romano J, Rodríguez MH, Pando V, Torres-Monzón JA, Alvarado-Delgado A, Lecona Valera AN, Ramos RA, Martínez-Barnetche J, Rodríguez MC |year=2011 |title=Conserved peptide sequences bind to actin and enolase on the surface of ''Plasmodium berghei'' ookinetes |journal=Parasitology |pmid=21816124 |doi=10.1017/S0031182011001296 |volume=138 |issue=11 |pages=1341–53}}</ref> |
|||
The myosin gene (MyoA) class XIV is essential for ookinete motility in the mosquito. Mutants placed under a different promotor active in the blood stages failed to complete their life cycle.<ref name=Siden-Kiamos2011>Siden-Kiamos I, Ganter M, Kunze A, Hliscs M, Steinbüchel M, Mendoza J, Sinden RE, Louis C, Matuschewski K (2011) Stage-specific depletion of Myosin A supports an essential role in motility of malarial ookinetes. Cell Microbiol {{doi|10.1111/j.1462-5822.2011.01686.x}}</ref> Disruption serine repeat antigen 5 blocks parasite inhibits egress of sporozoites from an oocyst.<ref name=Aly2005>{{cite journal |author=Aly AS, Matuschewski K |year=2005 |title=A malarial cysteine protease is necessary for Plasmodium sporozoite egress from oocysts |journal=J Exp Med |volume=202 |issue=2 |pages=225–230 |doi=10.1084/jem.20050545 |pmid=16027235 |pmc=2213010}}</ref> |
|||
Within the genome is encoded a protein phosphatase with a domain architecture known only otherwise to occur in [[Plantae]]: protein phosphatase containing kelch-like domains (PPKL).<ref name=Philip2012>Philip N, Vaikkinen HJ, Tetley L, Waters AP (2012) A unique kelch domain phosphatase in Plasmodium regulates ookinete morphology, motility and invasion" ''PLoS One'' 7(9) e44617</ref><ref name=Kutuzov2002>Kutuzov MA, Andreeva AV (2002) Protein Ser/Thr phosphatases with kelch-like repeat domains. Cell Signal 14: 745–750</ref> The kelch motif normally occurs as a series of four to seven repeats forming a beta propeller tertiary structure and may occur at either terminus of the protein. In ''Plasmodium'' PPKL has five full and a one half kelch domains with 5 inserts unique to Alveolates. This protein is produced in schizonts and female gametocytes and is maternally inherited. Absence of this protein results in leads to the development of a malformed, immotile, non infectious ookinetes with extended apical protrusions. This protein is localised at the ookinete apical tip. Secretion of microneme contents is unaffected. |
|||
Development of the motile and invasive ookinete within the mosquito midgut is dependent upon two NIMA-related kinases, NEK2 and NEK4.<ref name=Reininger2005>Reininger L, Billker O, Tewari R, Mukhopadhyay A, Fennell C, et al. (2005) A NIMA-related protein kinase is essential for completion of the sexual cycle of malaria parasites" ''J Biol Chem'' 280: 31957–31964</ref><ref name=Reininger2009>Reininger L, Tewari R, Fennell C, Holland Z, Goldring D, et al. (2009) An essential role for the ''Plasmodium'' Nek-2 Nima-related protein kinase in the sexual development of malaria parasites" ''J Biol Chem'' 284: 20858–20868</ref> |
|||
Two [[potassium]] channel have been identified in the genome.<ref name=Ellekvist2008>Ellekvist P, Maciel J, Mlambo G, Ricke CH, Colding H, Klaerke DA, Kumar N (2008) Critical role of a K+ channel in ''Plasmodium berghei'' transmission revealed by targeted gene disruption" ''Proc Natl Acad Sci USA'' 105(17) 6398-6402</ref> At least one of these is critical for the development of the ookinete in the mosquito. |
|||
The meiotic specific [[recombinase]] Dmc1 - a bacterial [[RecA]] homolog - appears to be essential for normal oocyst development in ''[[Plasmodium berghei]]''.<ref name=Mlambo2012>Mlambo G, Coppens I, Kumar N (2012) Aberrant sporogonic development of Dmc1 (a meiotic recombinase) deficient ''Plasmodium berghei'' parasites" ''PLoS One'' 7(12) e52480. {{DOI|10.1371/journal.pone.0052480}}</ref> |
|||
===Sporogony=== |
|||
Over the period of a 1–3 weeks, the [[oocyst]] grows to a size of tens to hundreds of micrometres. During this time, multiple nuclear divisions occur. After [[oocyst]] maturation is complete, the [[oocyst]] divides to form multiple haploid sporozoites. The number of sporozoiter produced per oocyst varies but the mean is ~500-700 per oocyst.<ref name=Vaughan1992>Vaughan JA, Noden BH, Beier JC (1992) Population dynamics of ''Plasmodium falciparum'' sporogony in laboratory-infected Anopheles gambiae. J Parasitol 78(4) 716-724</ref><ref name=Zollner2006>Zollner GE, Ponsa N, Garman GW, Poudel S, Bell JA, Sattabongkot J, Coleman RE, Vaughan JA (2006) Population dynamics of sporogony for ''Plasmodium vivax'' parasites from western Thailand developing within three species of colonized ''Anopheles'' mosquitoes. Malar J 5:68</ref> Immature sporozoites break through the [[oocyst]] wall into the haemolymph. The sporozoites then migrate to the salivary glands and complete their differentiation. Once mature, the sporozoites can proceed to infect a human host during a subsequent [[mosquito]] bite. |
|||
==Cell biology== |
|||
[[File:Plasmodium.png|right|thumb|200px| Diagram of the major organelles including the rhoptries, micronemes and polar rings near the apical end.]] |
|||
===Morphology=== |
|||
The nucleus, mitochondrion, apicoplast and the [[microtubule]]s of ''Plasmodium'' sporozoites are linked to the parasite [[pellicle (biology)|pellicle]] via long tethering proteins. The tethers originate from the inner membrane complex and are arranged in a periodic fashion following a 32 [[nanometer]] repeat. The tethers pass through a subpellicular structure that encompasses the entire parasite, probably as a network of membrane associated filaments.<ref name="Kudryashev2009">{{cite journal |author=Kudryashev M, Lepper S, Stanway R, ''et al''. |title=Positioning of large organelles by a membrane- associated cytoskeleton in Plasmodium sporozoites |journal=Cell. Microbiol. |volume=12 |issue=3 |pages=362–71 |year=2010 |month=March |pmid=19863555 |doi=10.1111/j.1462-5822.2009.01399.x}}</ref> |
|||
The volume of the nucleus of the asexual stages (~5 µm<sup>3</sup>) is approximately constant in size in the ring and trophozoite stages.<ref name=Moraes2013>Moraes CB, Dorval T, Contreras-Dominguez M, Dossin Fde M, Hansen MA, Genovesio A, Freitas-Junior LH (2013) Transcription sites are developmentally regulated during the asexual cycle of ''Plasmodium falciparum''. 8(2) e55539. {{DOI|10.1371/journal.pone.0055539}}</ref> The nucleus is divided into several subcompartments including the nucleolus and the nuclear periphery which appear to be involved in transcription control. |
|||
The pellicle is a structure unique to the Apicomplexa and is made up of four components: the plasma membrane, the inner membrane complex, the subpellicular network and the subpellicular microtubules.<ref name=Tremp2008>Tremp AZ, Khater EI, Dessens JT (2008) IMC1b is a putative membrane skeleton protein involved in cell shape, mechanical strength, motility, and infectivity of malaria ookinetes" ''J Biol Chem'' 283(41) 27604-27611</ref> The subpellicular network consists of a two-dimensional network of intermediate filaments located on the cytoplasmic side of the inner membrane complex and acts as a membrane skeleton. The proteins - the inner membrane complex proteins (IMCs) - that compose this structure are functional homologs of the [[articulin]]s, the membrane skeleton proteins of free-living protists. |
|||
The subpellicular network contains [[alveolin]]s - a group of proteins that determine the shape of the parasite.<ref name=Tremp2013>Tremp A, Carter V, Saeed S, Dessens JT (2013) Morphogenesis of ''Plasmodium'' zoites is uncoupled from tensile strength. Mol Microbiol {{DOI|10.1111/mmi.12297}}</ref> Another protein - G2 - which is structurally unrelated to alveolins appears to be involved in the organisation of the subpellicular microtubules. |
|||
===Cell division=== |
|||
Cell division occurs through a process known as schizogony. This is a type of mitotic division in which multiple rounds of nuclear divisions occur before the cytoplasm segments. |
|||
DNA synthesis begins in the relatively small trophozoites but nuclear subdivision, which leads to the formation of multinucleate cells, occurs only during schizogony. Whether or not any gap phases exist between each round of DNA synthesis and mitosis is unknown. Eventually, a schizont composed of 8–32 nuclei undergoes segmentation, which culminates with the formation of individual merozoites that burst from the erythrocyte into the blood stream. |
|||
==Cell Biology== |
|||
===Cell Division=== |
|||
Cell division occurs through a process known as schizogony. This is a type of mitotic division in which multiple rounds of nuclear divisions occur before the cytoplasm segments. |
|||
===Apical complex=== |
===Apical complex=== |
||
===Transport/Secretion=== |
|||
====Ions==== |
|||
The rhopteries appear to have subcompartments allowing for differential secretion during the life cycle.<ref name=Zuccala2012>Zuccala ES, Gout AM, Dekiwadia C, Marapana DS, Angrisano F, Turnbull L, Riglar DT, Rogers KL, Whitchurch CB, Ralph SA, Speed TP, Baum J (2012) Subcompartmentalisation of proteins in the rhoptries correlates with ordered events of erythrocyte invasion by the blood stage malaria parasite" ''PLoS One'' 7(9) e46160</ref> Two of these are known as the neck and the bulb.<ref name=Kemp2012>Kemp LE, Yamamoto M, Soldati-Favre D (2012) Subversion of host cellular functions by the apicomplexan parasites. FEMS Microbiol Rev {{DOI|10.1111/1574-6976.12013}}</ref> A number of rhoptry neck proteins are conserved between apicomplexan species and are involved in host cell invasion. Bulb proteins in contrast are less well conserved between the apicomplexa and most likely evolved for a particular lifestyle. In the majority of species studied to date, rhoptry content is involved in formation and maintenance of the parasitophorous vacuole. |
|||
===Parasitophorous Vacuole=== |
|||
Within a red blood cell, ''[[Plasmodium falciparum|P. falciparum]]'' resides inside the parasitophorous vacuole. This is formed during erythrocyte invasion. |
|||
;Rhoptery proteins |
|||
The proteins originating in the parasite pass through the membrane of the parasitophorous vacuole, and are transported to the cytoplasm or membrane of the erythrocyte.<ref name="gardner"/> This transport mechanism is largely unknown. |
|||
The rhoptery neck proteins (RONs) along with the micronemal AMA1 protein are important in the penetration of the erythrocyte.<ref name="Giovannini2011"/> The invasion mechanism while common to all Apicomplexa is unique and involves a tight interaction between the host cell and the parasite surfaces called the moving junction. The moving junction, which is the anchoring structure for the invasion process, is formed by secretion of a macromolecular complex (proteins RON-2, -4, -5, -8), derived from the rhoptries, into the host cell membrane. |
|||
The mechanisms involved in this process are still being elucidated. The protein RON8 appears to be central to the binding of parasite to the erythrocyte surface.<ref name="Straub2011">{{cite journal |author=Straub KW, Peng ED, Hajagos BE, Tyler JS, Bradley PJ |year=2011 |title=The moving junction protein RON8 facilitates firm attachment and host cell invasion in ''Toxoplasma gondii'' |journal=PLoS Pathog |volume=7 |issue=3 |page=e1002007 |doi=10.1371/journal.ppat.1002007 |editor1-last=Striepen |editor1-first=Boris}}</ref> Apical Sushi Protein and Rhoptry Neck protein 2 are released early following the formation of the tight junction between the merozoite and the erythrocyte. The rhoptry protein PFF0645c is released only after invasion is complete. |
|||
The ''P. falciparum'' apical sushi protein is the homolog of the ''P. vivax'' RON1 protein.<ref name=Moreno-Perez2011>{{cite journal |author=Moreno-Perez DA, Montenegro M, Patarroyo ME, Patarroyo MA |title=Identification, characterization and antigenicity of the ''Plasmodium vivax'' rhoptry neck protein 1 (PvRON1) |journal=Malar. J. |volume=10 |page=314 |year=2011 |pmid=22024312 |pmc=3215230 |doi=10.1186/1475-2875-10-314}}</ref> |
|||
The rhoptry protein 2 of ''[[Plasmodium vivax]]'' has been cloned.<ref name=Wang2012>Wang B, Lu F, Cheng Y, Li J, Ito D, Sattabongkot J, Tsuboi T, Han ET (2012) Identification and characterization of the ''Plasmodium falciparum'' RhopH2 ortholog in Plasmodium vivax. Parasitol Res</ref> The 1,369 amino acid protein is encoded PVX_099930 gene. The gene has nine introns and the protein contains a signal peptide at its N-terminus and 12 cysteines predominantly in its C-terminal half. It is localized in one of the apical organelles of the merozoite, the rhoptry, and the localization pattern is similar to its homolog in ''P. falciparum''. |
|||
RON2 is inserted as an integral membrane protein in the host cell.<ref name=Lamarque2011>Lamarque M, Besteiro S, Papoin J, Roques M, Vulliez-Le Normand B, Morlon-Guyot J, Dubremetz JF, Fauquenoy S, Tomavo S, Faber BW, Kocken CH, Thomas AW, Boulanger MJ, Bentley GA, Lebrun M (2011) The RON2-AMA1 interaction is a critical step in moving junction-dependent invasion by apicomplexan parasites" ''PLoS Pathog'' 7(2) e1001276. {{DOI|10.1371/journal.ppat.1001276}}</ref> |
|||
The residues of the RON2 protein that bind to the AMA-1 protein have been identified.<ref name=Srinivasan2011>{{cite journal |author=Srinivasan P, Beatty WL, Diouf A, Herrera R, Ambroggio X, Moch JK, Tyler JS, Narum DL, Pierce SK, Boothroyd JC, Haynes JD, Miller LH |year=2011 |title=Binding of ''Plasmodium'' merozoite proteins RON2 and AMA1 triggers commitment to invasion |journal=Proc Natl Acad Sci U S A |doi=10.1073/pnas.1110303108 |volume=108 |issue=32 |pages=13275–80 |pmid=21788485 |pmc=3156155}}</ref> It also appears that the formation of the junction and parasitophorous vacuole are molecularly distinct steps in the invasion process. Positive diversifying selection appears to have acted in the RON2 protein of ''[[Plasmodium vivax]]''.<ref name=Tang2012>Tang J, Dai Y, Zhang H, Culleton RL, Liu Y, Zhao S, Wang X, Guan X, Kaneko O, Zhu Y (2012) Positive diversifying selection on Plasmodium vivax RON2 protein. Parasitology</ref> |
|||
In ''Plasmodium berghei'' RON4 is required for sporozoite invasion of hepatocytes.<ref name=Giovannini2011>Giovannini D, Späth S, Lacroix C, Perazzi A, Bargieri D, Lagal V, Lebugle C, Combe A, Thiberge S, Baldacci P, Tardieux I, Ménard R (2011) Independent roles of apical membrane antigen 1 and rhoptry neck proteins during host cell invasion by apicomplexa. Cell Host Microbe 10(6) 591-602. {{DOI|10.1016/j.chom.2011.10.012}}</ref> |
|||
RhopH2 is transcribed and localised to the endoplasmic reticulum of the trophozoites by 28 hours post invasion.<ref name=Ghoneim2013>Ghoneim AM (2013) Trafficking of ''Plasmodium falciparum'' chimeric rhoptry protein with Brefeldin A. Folia Parasitol (Praha) 60(1) 75-78</ref> This pathway is Brefeldin A-sensitive. By 32 hours the protein is distributed in the schizonts' cytoplasm but not in the parasitophorous vacuole. |
|||
A protein - Armadillo Repeats-Only - has been localised to the cytosolic face of the [[rhoptry|rhoptries]].<ref name=Cabrera2010>Cabrera A, Herrmann S, Warszta D, Santos JM, John Peter AT, Kono M, Debrouver S, Jacobs T, Spielmann T, Ungermann C, Soldati-Favre D, Gilberger TW (2012) Dissection of minimal sequence requirements for rhoptry membrane targeting in the malaria parasite" ''Traffic'' 9999(999A). {{DOI|10.1111/j.1600-0854.2012.01394.x}}</ref> A putative signal sequence in the first 20 amino acids has also been identified. |
|||
A number of heparin binding proteins are present in the rhopteries.<ref name=Zhang2013>Zhang Y, Jiang N, Lu H, Hou N, Piao X, Cai P, Yin J, Wahlgren M, Chen Q (2013) Proteomic analysis of Plasmodium falciparum schizonts reveals heparin-binding merozoite proteins. J Proteome Res</ref> [[Heparin]] like molecules bound to the surface of the erythrocyte appear to be important in the adhesion process which also involves the merozoite surface protein 1.<ref name="Boyle2010"/> |
|||
A protein S-acyl transferase with a DHHC-cysteine-rich domain is present in the rhoptry.<ref name="Frénal2013">Frénal K, Tay CL, Mueller C, Bushell ES, Jia Y, Graindorge A, Billker O, Rayner JC, Soldati-Favre D (2013) Global analysis of apicomplexan protein S-acyl transferases reveals an enzyme essential for invasion. Traffic {{DOI|10.1111/tra.12081}}</ref> This enzyme may play a role in the apical positioning of the rhoptry organelles. |
|||
;Micronemal proteins |
|||
Apical membrane antigen-1 (AMA-1) - the product of the Pf''ama1'' gene - is a surface exposed protein that plays a role in erythrocyte invasion. It is shed from the parasite surface predominantly via the action of the protease Sub2<ref name=Olivieri2011>{{cite journal |author=Olivieri A, Collins CR, Hackett F, Withers-Martinez C, Marshall J, Flynn HR, Skehel JM, Blackman MJ |year=2011 |title=Juxtamembrane shedding of ''Plasmodium falciparum'' AMA1 is sequence independent and essential, and helps evade invasion-inhibitory antibodies |journal=PLoS Pathog |volume=7 |issue=12 |page=e1002448 |doi=10.1371/journal.ppat.1002448 |editor1-last=Carruthers |editor1-first=Vern B}}</ref> Using gene deletion mutants AMA-1 has been shown to be a non essential protein.<ref name=Bargieri2013>Bargieri DY, Andenmatten N, Lagal V, Thiberge S, Whitelaw JA, Tardieux I, Meissner M, Ménard R (2013) Apical membrane antigen 1 mediates apicomplexan parasite attachment but is dispensable for host cell invasion. Nat Commun 4:2552. {{DOI|10.1038/ncomms3552}}</ref> While attached to the surface of the parasite AMA-1 can bind to RON2 - a protein that is inserted by the parasite into the erythrocyte membrane. This binding seems to be involved in the invasion process. |
|||
The receptor binding site of AMA-1 comprises the hydrophobic groove and a region that becomes exposed by displacement of the flexible domain II loop.<ref name=Vulliez-Le2012>{{cite journal |last1=Vulliez-Le Normand |first1=B |last2=Tonkin |first2=ML |last3=Lamarque |first3=MH |last4=Langer |first4=S |last5=Hoos |first5=S |last6=Roques |first6=M |last7=Saul |first7=FA |last8=Faber |first8=BW |last9=Bentley |first9=GA ''et al.'' |year=2012 |month=Jun |title=Structural and functional insights into the malaria parasite moving junction complex |journal=PLoS Pathog |volume=8 |issue=6 |page=e1002755 |doi=10.1371/journal.ppat.1002755 |editor1-last=Phillips |editor1-first=Meg |last10=Boulanger |first10=Martin J. |last11=Lebrun |first11=Maryse}}</ref> |
|||
Sub2 is released from the micronemes and can also act on the MSP1/6/7 complex and PTRAMP - another micronemal protein. Sub2 appears to be an essential gene. |
|||
;Proteins whose location is unclear |
|||
Several proteins are involved in the binding of the sporozoite to the various tissues it attaches to. [[Attenuator (genetics)|TRAP]], [[S6 (gene)|S6]] and [[TLP (gene)|TLP]] have been implicated in these processes.<ref name="Hegge2010">{{cite journal |author=Hegge S, Münter S, Steinbüchel M, Heiss K, Engel U, Matuschewski K, Frischknecht F |year=2010 |title=Multistep adhesion of ''Plasmodium'' sporozoites |journal=FASEB J. |doi=10.1096/fj.09-148700 |volume=24 |issue=7 |pages=2222–34 |pmid=20159960}}</ref> |
|||
===Maurer's clefts=== |
|||
In 1902 the German physician [[Georg Maurer]] discovered an unusual staining pattern in the cytoplasm of erythrocytes infected with ''P. falciparum''. These structures were subsequently named Maurer's clefts. These consist of a convoluted set of membranes that lie within the erythrocyte's cytoplasm and appear to be involved in secrection from the erythrocyte.<ref name=Wickert2004>Wickert H, Göttler W, Krohne G, Lanzer M (2004) Maurer's cleft organization in the cytoplasm of ''Plasmodium falciparum''-infected erythrocytes: new insights from three-dimensional reconstruction of serial ultrathin sections. Eur J Cell Biol 83(10) 567-582</ref> They are known to have proteins of parasite origin within them including the Maurer's cleft two transmembrane proteins (PfMC-2TM)<ref name="Tsarukyanova2009">{{cite journal |author=Tsarukyanova I, Drazba JA, Fujioka H, Yadav SP, Sam-Yellowe TY |year=2009 |title=Proteins of the ''Plasmodium falciparum'' two transmembrane Maurer's cleft protein family, PfMC-2TM, and the 130 kDa Maurer's cleft protein define different domains of the infected erythrocyte intramembranous network |journal=Parasitol Res |volume=104 |issue=4 |pages=875–891 |doi=10.1007/s00436-008-1270-3 |pmid=19130087}}</ref> The clefts appear to originate from vacuoles budding off them the parasitophorous vacuole membrane which then diffuse within the erythrocyte cytoplasm before taking up residence at the cell periphery.<ref name="Spycher2006">{{cite journal |author=Spycher C, Rug M, Klonis N, Ferguson DJ, Cowman AF, Beck HP, Tilley L |year=2006 |title=Genesis of and Trafficking to the Maurer's Clefts of Plasmodium falciparum-Infected Erythrocytes |journal=Mol Cell Biol |volume=26 |issue=11 |pages=4074–4085 |doi=10.1128/MCB.00095-06 |pmid=16705161 |pmc=1489082}}</ref> |
|||
Another protein associated with these structure is skeleton-binding protein 1 (SBP1). This protein is involved in transport of the ''var'' gene protein, PfEMP1 (erythrocyte membrane protein 1) to the erythrocyte surface.<ref name="Cooke2006">{{cite journal |author=Cooke BM, Buckingham DW, Glenister FK, Fernandez KM, Bannister LH, Marti M, Mohandas N, Coppel RL |year=2006 |title=A Maurer's cleft–associated protein is essential for expression of the major malaria virulence antigen on the surface of infected red blood cells |journal=J Cell Biol |volume=172 |issue=6 |pages=899–908 |doi=10.1083/jcb.200509122 |pmid=16520384 |pmc=2063733}}</ref> |
|||
Other proteins associated with these structures include membrane associated histidine rich protein 1 and ring exported protein 1 and 2.<ref name=Spycher2003>Spycher C, Klonis N, Spielmann T, Kump E, Steiger S, ''et al'' (2003) MAHRP-1, a novel ''Plasmodium falciparum'' histidine-rich protein, binds ferriprotoporphyrin IX and localizes to the Maurer's clefts" ''J Biol Chem'' 278: 35373–35383</ref><ref name=Hawthorne2004>Hawthorne PL, Trenholme KR, Skinner-Adams TS, Spielmann T, Fischer K, ''et al'' A novel ''Plasmodium falciparum'' ring stage protein, REX, is located in Maurer's clefts. Mol Biochem Parasitol 136: 181–189</ref> |
|||
Mutations in the ring exported protein 1 (Rex 1), a protein normally found in Maurer's clefts, reduces transport of the ''var'' gene products to the erythrocyte surface.<ref name=Dixon2011>Dixon MW, Kenny S, McMillan PJ, Hanssen E, Trenholme KR, Gardiner DL, Tilley L (2011) Genetic ablation of a Maurer's cleft protein prevents assembly of the ''Plasmodium falciparum'' virulence complex. Mol Microbiol {{doi|10.1111/j.1365-2958.2011.07740.x}}</ref> |
|||
The erythrocyte protein [[ankyrin]] is found in these structures.<ref name=Atkinson2986>Atkinson CT, Aikawa M, Perry G, Fujino T, Bennett V, Davidson EA, Howard RJ (1988) Ultrastructural localization of erythrocyte cytoskeletal and integral membrane proteins in Plasmodium falciparum-infected erythrocytes. Eur J Cell Biol 45(2) 192-199</ref> |
|||
The parasite generates a host derived actin cytoskeleton within the cytoplasm of the erythrocytes that connects the Maurer's clefts with the host cell membrane and to which transport vesicles are attached.<ref name=Cyrklaff2012>Cyrklaff M, Sanchez CP, Kilian N, Bisseye C, Simpore J, Frischknecht F, Lanzer M (2012) Hemoglobins S and C interfere with actin remodeling in ''Plasmodium falciparum''-infected erythrocytes" ''Science'' 334(6060) 1283-1286</ref> Hemoglobin oxidation products which are enriched in hemoglobin S and C containing erythrocytes inhibit actin polymerization. This may account for their protective role in malaria. |
|||
The protein trophozoite exported protein 1 (PFF0165c) is located within the clefts.<ref name=Kulangara2012>Kulangara C, Luedin S, Dietz O, Rusch S, Frank G, Mueller D, Moser M, Kajava AV, Corradin G, Beck HP, Felger I (2012) Cell biological characterization of the malaria vaccine candidate trophozoite exported protein 1" ''PLoS One'' 7(10) e46112. {{DOI|10.1371/journal.pone.0046112}}</ref> The protein's N-terminal region is intrinsically unstructured but it also has a coiled coil domain. It appears to lack export motifs such as PEXEL, signal sequence/anchor or a transmembrane domain. Transport of this protein to the clefts is sensitive to inhibition by [[Brefeldin A]]. This is normally associated with proteins that are have co-translation translocation into the endoplasmic reticulum or posttranslational insertion into the endoplasmic reticulum followed by vesicular transport from the endoplasmic reticulum via Golgi apperatus to the cell surface. |
|||
Mutations in REX1 and Pf332 proteins result in distortion of Maurer's clefts morphology suggesting that they play a role in its structure.<ref name=Hanssen2008>Hanssen E, Hawthorne P, Dixon MW, Trenholme KR, McMillan PJ ''et al'' (2008) Targeted mutagenesis of the ring-exported protein-1 of ''Plasmodium falciparum'' disrupts the architecture of Maurer's cleft organelles" ''Mol Microbiol'' 69: 938–953</ref><ref name=Glenister2009>Glenister FK, Fernandez KM, Kats LM, Hanssen E, Mohandas N ''et al'' (2009) Functional alteration of red blood cells by a megadalton protein of ''Plasmodium falciparum''" ''Blood'' 113: 919–928</ref> |
|||
The actin network exerts skeletal functions by anchoring the Maurer's clefts within the erythrocyte cytoplasm and restrains the Brownian motion of this organelle.<ref name=Kilian2012>Kilian N, Dittmer M, Cyrklaff M, Ouermi D, Bisseye C, Simpore J, Frischknecht F, Sanchez CP, Lanzer M (2012) Hemoglobin S and C affect the motion of Maurer's clefts in ''P. falciparum''-infected erythrocytes. Cell Microbiol {{DOI|10.1111/cmi.12102}}</ref> Haemoglobin S and C appear to interfere with this organisation. |
|||
The protein Surfin 4.1 is found in the clefts.<ref name=Zhu2012>Zhu X, Yahata K, Alexandre JS, Tsuboi T, Kaneko O (2012) The N-terminal segment of ''Plasmodium falciparum'' SURFIN(4.1) is required for its trafficking to the red blood cell cytosol through the endoplasmic reticulum. Parasitol Int pii: S1383-5769(12)00166-3. {{DOI|10.1016/j.parint.2012.12.006}}</ref> |
|||
The membrane associated histidine rich protein 2 is attached to Maurer's clefts.<ref name=McMillan2013>McMillan PJ, Millet C, Batinovic S, Maiorca M, Hanssen E, Kenny S, Muhle RA, Melcher M, Fidock DA, Smith JD, Dixon MW, Tilley L (2013) Spatial and temporal mapping of the PfEMP1 export pathway in ''Plasmodium falciparum''. Cell Microbiol {{DOI|10.1111/cmi.12125}}</ref> |
|||
There are three multigene families organized into 9 highly conserved clusters with the Pfmc-2tm genes in the subtelomeric regions of the chromosomes.<ref name=Mbengue2013>Mbengue A, Audiger N, Vialla E, Dubremetz JF, Braun-Breton C (2013) Novel ''Plasmodium falciparum'' Maurer's clefts protein families implicated in the release of infectious merozoites. Mol Microbiol {{DOI|10.1111/mmi.12193}}</ref> These genes are expressed at early trophozoite stages. Like the PfMC-2TM proteins, the PfEPF1, 3 and 4 proteins encoded by these families are exported to the Maurer's clefts, as peripheral or integral proteins of the Maurer's cleft membrane and largely exposed to the red cell cytosolic face of this membrane. |
|||
Within the PfMC-2TM proteins there is a conserved domain MC-TYR.<ref name=Frech2013>Frech C, Chen N (2013) Variant surface antigens of malaria parasites: functional and evolutionary insights from comparative gene family classification and analysis" ''BMC Genomics'' 14(1) 427</ref> |
|||
===Dense granules=== |
|||
These are small vacuoles that can be seen on electron microscopy. They bind [[osmium]] and appear dark on the images. Little is known about these organelles but they are thought to play a role in the maintenance of the parasitophorous vacuole. One protein - p377 - has been localised to these organelles. Disruption of this protein reduces osmiophilic body formation, and leads to a marked decrease in female fitness and impaired infectivity to mosquitoes.<ref name=Hayton2008>Hayton K, Templeton TJ (2008) Osmiophilic bodies and the odd organelles of alveolates" ''Mol Microbiol'' 67(2) 236-240</ref> |
|||
A protein - the ring membrane antigen (RIMA) - has been localised to the dense granules.<ref name=Trager1992>Trager W, Rozario C, Shio H, Williams J, Perkins ME (1992) Transfer of a dense granule protein of ''Plasmodium falciparum'' to the membrane of ring stages and isolation of dense granules" ''Infect Immun'' 60(11) 4656-4661</ref> The protein's molecular weight is 14 kiloDaltons and it is synthesized late in schizogony. At the late schizont stage this protein is distributed diffusely throughout the intracellular schizont. During the segmenter stage it is then localized to the dense granules. |
|||
===Parasitophorous vacuole=== |
|||
Within a red blood cell, ''[[Plasmodium falciparum|P. falciparum]]'' resides inside the parasitophorous vacuole. This is formed during erythrocyte invasion. |
|||
The proteins originating in the parasite pass through the membrane of the parasitophorous vacuole and are transported to the cytoplasm or membrane of the erythrocyte.<ref name="gardner"/> Although this transport mechanism is largely unknown some details have been elucidated.<ref name="AbuBakar2010">{{cite journal |author=Abu Bakar N, Klonis N, Hanssen E, Chan C, Tilley L |year=2010 |title=Digestive-vacuole genesis and endocytic processes in the early intraerythrocytic stages of ''Plasmodium falciparum'' |journal=J Cell Sci.}}</ref> Ingestion of the erythrocyte cytoplasm begins in mid-ring-stage parasites. Host [[cytoplasm]] is internalised via [[cytostome]]-derived invaginations and then concentrated into several acidified peripheral structures. [[Haemoglobin]] digestion and [[hemozoin|haemozoin]] formation occur within these [[Vesicle (biology)|vesicle]]s. The ring-stage parasites can adopt a deeply invaginated cup shape, but they do not take up haemoglobin via [[macropinocytosis]]. As the parasite matures the haemozoin containing compartments coalesce to form a single acidic digestive vacuole (pH 4.5 - 5.5) that is then fed by haemoglobin containing vesicles. Some haemoglobin degradation also occurs in compartments outside the [[digestive vacuole]]. |
|||
The enzyme [[phosphatidyl-inositol-3-kinase]] (PI3K) has been implicated in this process.<ref name="Vaid2010">{{cite journal |author=Vaid A, Ranjan R, Smythe WA, Hoppe HC, Sharma P |year=2010 |title=PfPI3K, a Phosphatidylinositol-3 kinase from ''Plasmodium falciparum'', is exported to the host erythrocyte and is involved in hemoglobin trafficking |journal=Blood |doi=10.1182/blood-2009-08-238972 |volume=115 |issue=12 |pages=2500–7 |pmid=20093402 |pmc=2918364}}</ref> PI3K is located in vesicular compartments near the membrane and in the digestive vacuole and is involved in endocytosis from the host and trafficking of hemoglobin in the parasite. Its inhibition with [[wortmannin]] or [[LY294002]] results in entrapment of hemoglobin in vesicles within the parasite cytoplasm preventing its transport to the [[food vacuole|digestive vacuole]]. |
|||
The pH of the digestive vacuole is maintained by a V-type H(+)-ATPase. |
|||
A signal sequence at the N terminal of proteins targeted to the arasitophorous vacuole has been identified.<ref name="Eksi2011">{{cite journal |author=Eksi S, Williamson KC |year=2011 |title=Protein targeting to the parasitophorous vacuole membrane of ''Plasmodium falciparum.'' |journal=[[Eukaryotic Cell]] |doi=10.1128/EC.00008-11 |volume=10 |issue=6 |pages=744}}</ref> The signal appears to reside in the 55 amino acids of the N terminal of the protein. There may be a retention signal at the C terminal. |
|||
The micronemal protease [[Plasmodium ROM1|ROM1]] appears to be essential for proper parasitophorous vacuole modification to allow parasite development.<ref name=Vera2011>{{cite journal |author=Vera IM, Beatty WL, Sinnis P, Kim K |year=2011 |title=Plasmodium'' protease ROM1 is important for proper formation of the parasitophorous vacuole |journal=PLoS Pathog |volume=7 |issue=9 |page=e1002197 |doi=10.1371/journal.ppat.1002197 |editor1-last=Deitsch |editor1-first=Kirk}}</ref> This protease is able to cleave the proteins AMA1 and MAEBL. |
|||
[[Uric acid]] precipitates are present in the cytoplasm of the parasitophorous vacuole.<ref name=van_de_Hoef2013>van de Hoef DL, Coppens I, Holowka T, Ben Mamoun C, Branch O, Rodriguez A (2013) ''Plasmodium falciparum''-derived uric acid precipitates induce maturation of dendritic cells" ''PLoS One'' 8(2) e55584. {{DOI|10.1371/journal.pone.0055584}}</ref> These are released when the merozoites are released. Uric acid is highly inflammatory and can cause maturation of dendritic cells. |
|||
The protein PFA0210c is a member of the START domain family which is involved in the transport of phospholipids, ceramide or fatty acids between membranes.<ref name=vanOoij2013>van Ooij C, Withers-Martinez C, Ringel A, Cockcroft S, Haldar K, Blackman MJ (2013) Identification of a ''Plasmodium falciparum'' phospholipid transfer protein. J Biol Chem</ref> It associates with membranes in infected erythrocytes at mature stages of intracellular parasite growth. It is present in the parasitophorous vacuole during growth and is later recruited to organelles in the parasite. |
|||
===Apicoplast=== |
===Apicoplast=== |
||
''[[Plasmodium falciparum]]'', and other members of the [[apicomplexa]] [[phylum]], contain an [[organelle]] called the [[apicoplast]].<ref name="gardner"/> The [[apicoplast]] is an essential [[plastid]], homologous to a [[chloroplast]], although the [[apicoplast]] is not [[photosynthetic]]. Evolutionarily, it is thought to have have derived through secondary [[endosymbiosis]]. |
|||
''[[Plasmodium falciparum]]'', and most other members of the [[phylum]] [[Apicomplexa]], contain an [[organelle]] termed an [[apicoplast]].<ref name="gardner"/> It was first discovered in 1996.<ref>Biot C, Botté CY, Dubar F, Maréchal E (2002) Targeting malaria parasite at the level of apicoplast: an update. Med Sci (Paris) 28(2) 163-171. {{DOI|10.1051/medsci/2012282014}}</ref> The [[apicoplast]] is an [[essential (biology)|essential]] [[plastid]], homologous to a [[chloroplast]], although the [[apicoplast]] itself lacks any [[photosynthetic]] function. Evolutionarily it is thought to have been derived through secondary [[endosymbiosis]]. As humans do not harbor apicoplasts, this organelle and its constituents are seen as a possible target for antimalarial drugs. |
|||
The function of the [[apicoplast]] remains to be fully determined, but it appears to be involved in the metabolism of [[fatty acids]], [[isoprenoids]], and [[heme]].<ref name="gardner"/> |
|||
The apicoplast varies in size during the life cycle from ~0.5 µm × 0.15 µm in the merozoite to 1.6 µm × 0.35 µm in the trophozoites.<ref name=Hopkins1999>Hopkins J, Fowler R, Krishna S, Wilson I, Mitchell G, Bannister L (1999) The plastid in ''Plasmodium falciparum'' asexual blood stages: a three-dimensional ultrastructural analysis" ''Protist'' 150(3) 283-295</ref> Only one copy is present until it replicates in late schizonts. The apicoplast always adheres to the mitochondrion, along its whole length in merozoites and early rings but only at one end in later stages. Regions of the apicoplast are also closely related to the pigment vacuole, nuclear membrane and endoplasmic reticulum. In merozoites the plastid is anchored to a band of 2-3 subpellicular microtubules. |
|||
The [[apicoplast]] contains a 35-kb [[genome]], which encodes for 30 [[proteins]]. Other, nuclear-encoded, [[proteins]] are transported into the [[apicoplast]] using a specific [[signal peptide]]. It is estimated that 551, or roughly 10%, of the predicted nuclear-encoded [[proteins]] are targeted to the [[apicoplast]].<ref name="gardner"/> |
|||
The apicoplast has four membranes is normally located between the nucleus and the rhoptries.<ref name=Lemgruber2013>Lemgruber L, Kudryashev M, Dekiwadia C, Riglar DT, Baum J, Stahlberg H, Ralph SA, Frischknecht F (2013) Cryo-electron tomography reveals four-membrane architecture of the ''Plasmodium'' apicoplast. Malar J 12(1) 25</ref> Its matrix contains ribosome sized particles and membranous whorls. The gap between the second and third membranes is frequently larger than between the other membranes. The interior matrix of the apicoplast contains ribosome-like granules and a network of fine branched filaments. |
|||
As humans do not harbor apicoplasts, this organelle and its constituents are seen as a possible target for antimalarial drugs. |
|||
It contains a 35-kb [[genome]], which encodes for 30 [[proteins]]. The genome of this organelle has now been sequenced for several species.<ref name=Arisue2012>Arisue N, Hashimoto T, Mitsui H, Palacpac NM, Kaneko A, Kawai S, Hasegawa M, Tanabe K, Horii T (2012) The Plasmodium apicoplast genome: conserved structure and close relationship of P. ovale to rodent malaria parasites. Mol Biol Evol</ref> It appears to be conserved and to encode ~30 genes in all species examined. |
|||
==Genome== |
|||
The genome of ''[[Plasmodium falciparum]]'' (clone 3D7) was fully sequenced in 2002. <ref name="gardner"/>. The parasite has a 23 megabase genome, divided into 14 [[chromosomes]].<ref name="gardner"/> The genome codes for approximately 5,300 genes. About 60% of the putative proteins have little or no similarity to proteins in other organisms, and thus currently have no functional assignment.<ref name="gardner">{{cite journal |
|||
| last = Gardner |
|||
| first = Malcom |
|||
| authorlink = |
|||
| coauthors = |
|||
| title = Genome sequence of the human malaria parasite ''Plasmodium falciparum'' |
|||
| journal = Nature |
|||
| volume = 419 |
|||
| issue = |
|||
| pages = 498–511 |
|||
| publisher = |
|||
| location = |
|||
| date = 3 October 2002 |
|||
| url = http://www.nature.com/nature/journal/v419/n6906/full/nature01097.html |
|||
| doi =10.1038/nature01097 |
|||
| id = |
|||
| accessdate = }}</ref> It is estimated 52.6% of the genome is a coding region, with 53.9% of the putative genes containing at least one intron. <ref name="gardner"/> |
|||
The plastid genome replicates at the late trophozoite stage of the parasite intraerythocytic cycle. It proceeds predominantly via a D-loop/bi-directional ''ori'' mechanism with replication ''ori'' localized within the inverted repeat region. The process of replication involves a nuclear-encoded [[DnaJ]] homolog that binds to the ''ori'' site.<ref name="Kumar2010">{{cite journal |author=Kumar A, Tanveer A, Biswas S, Ram ER, Gupta A, Kumar B, Habib S |year=2010 |title=Nuclear-encoded DnaJ homolog of Plasmodium falciparum interacts with replication ori of the apicoplast genome |journal=Mol. Microbiol. |doi=10.1111/j.1365-2958.2009.07033.x |volume=75 |issue=4 |pages=942–56 |pmid=20487289}}</ref> |
|||
===Haploid/Diploid=== |
|||
It is [[haploid]] during nearly all stages of its life-cycle, except for a brief period after fertilization when it is diploid from the [[ookinete]] to sporogenic stages within the [[mosquito]] gut. |
|||
The [[DNA polymerase]] involved in the replication of its genome is Pfprex (Klenow-like polymerase). This enzyme has been cloned, expressed and purified.<ref name=Kennedy2011>{{cite journal |author=Kennedy SR, Chen CY, Schmitt MW, Bower CN, Loeb LA |year=2011 |title=The biochemistry and fidelity of synthesis by the apicoplast genome replication DNA polymerase Pfprex from the malaria parasite ''Plasmodium falciparum'' |journal=J Mol Biol |doi=10.1016/j.jmb.2011.04.071 |volume=410 |pages=27–38 |pmid=21570407 |issue=1 |pmc=3117635}}</ref> The enzymes is relatively error prone and shows a bias toward T->C mutations. |
|||
===AT Richness=== |
|||
The ''[[Plasmodium falciparum|P. falciparum]]'' genome has an AT content of roughly 80.6%.<ref name="gardner"/> Within the intron and intergenic regions, this AT composition rises to roughly 90%. The putative exons contain an AT content of 76.3%. The parasite's AT content is very high in comparison to other organisms. For example, the entire genomes of ''[[Saccharomyces cerevisiae]]'' and ''[[Arabidopsis thaliana]]'' have AT contents of 62% and 65%, respectively.<ref name="gardner"/> |
|||
Other nuclear encoded [[proteins]] are transported into the [[apicoplast]]. Transport into the apicoplast are not well understood. These proteins has a signal in the N terminal but unlike many other organisms this appears to be a disordered chain rather than a conserved sequence.<ref name=Gallagher2011>Gallagher JR, Matthews KA, Prigge ST (2011) ''P. falciparum'' apicoplast transit peptides are unstructured ''in vitro'' and during apicoplast import. Traffic {{doi|10.1111/j.1600-0854.2011.01232.x}}</ref> It was thought that a specific [[signal peptide]] was responsible for this targeting <ref name="gardner"/> and it was estimated that 551, or roughly 10%, of the predicted nuclear-encoded [[proteins]] are targeted to the [[apicoplast]]. This hypothesis now appears to be incorrect. It appears that a relative enrichment within the protein of positively charged amino acid residues ([[Arginine]], [[Histidine]], [[Lysine]]) particularly at the N terminal of the protein may be sufficient to target the protein to the apicomplast.<ref name=Tonkin2006>Tonkin CJ, Roos DS, McFadden GI (2006) N-terminal positively charged amino acids, but not their exact position, are important for apicoplast transit peptide fidelity in ''Toxoplasma gondii''. Mol Biochem Parasitol 150(2) 192-200</ref> |
|||
===Promoters=== |
|||
===Subtelomeric regions=== |
|||
The subtelomeric regions of ''[[Plasmodium falciparum|P. falciparum]]'' chromosomes show a high degree of conservation within the genome, and contain significant amounts of repeated structure. <ref name="gardner"/> These conserved regions can be divided into five subtelomeric blocks. The blocks contain tandem repeats in addition to non-repetitive regions. |
|||
The biosynthesis of this organelle is not well understood. Phosphatidylinositol 3-monophosphate has been shown to be involved in its biosynthesis in the apicomplexian ''[[Toxoplasma gondii]]''.<ref name="Tawk2011">Tawk L, Dubremetz JF, Montcourrier P, Chicanne G, Merezegue F, Richard V, Payrastre B, Meissner M, Vial HJ, Roy C, Wengelnik K, Lebrun M (2011) Phosphatidylinositol 3-monophosphate is involved in toxoplasma apicoplast biogenesis. PloS Pathog. 7(2) e1001286</ref> It seems likely that this enzyme is involved in the formation of this organelle in the ''Plasmodium'' species also. |
|||
Many genes involved in antigenic variation are located in the subtelomeric regions of the chromosomes. These are divided into the ''var'', ''rif'', and ''stevor'' families. Within the genome, there exist 59 ''var'', 149 ''rif'', and 28 ''stevor'' genes, along with multiple pseudogenes and truncations.<ref name="gardner"/> |
|||
This organelle appears to be essential in the liver stages.<ref name=Biot2012>Biot C, Botté CY, Dubar F, Maréchal E (2012) Targeting malaria parasite at the level of apicoplast: an update. Med Sci (Paris) 28(2) 163-171</ref> |
|||
==Transcriptome== |
|||
[[Image:Pftranscriptome.jpg|thumb|Plaseogram of ''[[Plasmodium falciparum]]'' intraerythrocytic development cycle transcriptome<ref name="bozdech"/>]] |
|||
A transcriptome analysis has been conducted on the intraerythrocytic development cycle of ''[[Plasmodium falciparum|P. falciparum]]''.<ref name="bozdech">{{cite journal |
|||
| last = Bozdech |
|||
| first = Zbynek |
|||
| authorlink = |
|||
| coauthors = |
|||
| title = The Transcriptome of the Intraerythrocytic Developmental Cycle of Plasmodium falciparum |
|||
| journal = PLoS Biology |
|||
| volume = 1 |
|||
| issue = 1 |
|||
| pages = |
|||
| publisher = |
|||
| location = |
|||
| date = August 18, 2003 |
|||
| url = http://biology.plosjournals.org/perlserv/?request=get-document&doi=10.1371%2Fjournal.pbio.0000005 |
|||
| doi = |
|||
| id = |
|||
| accessdate = }}</ref> Roughly 60% of the genome is transcriptionally active during this portion of the parasite's life cycle. Whereas many genes appear to have stable mRNA levels throughout the cycle, many of the genes are transcriptionally regulated in a continuous cascade. |
|||
The functions of this organelle remains to be fully determined but it appears to be involved in the metabolism of [[fatty acids]], [[isoprenoids]] and [[heme]].<ref name="gardner"/> There are two pathways for protein lipoylation in ''Plasmodium'' - one in the mitochondrion and the other in the apicoplast. The apicoplast pathway is not found in the vertebrate host and relies on ''de novo'' [[lipoic acid]] synthesis.<ref name=Deschermeier2011>Deschermeier C, Hecht LS, Bach F, Rützel K, Stanway RR, Nagel A, Seeber F, Heussler VT (2011) Mitochondrial lipoic acid scavenging is essential for ''Plasmodium berghei'' liver stage development. Cell Microbiol {{doi|10.1111/j.1462-5822.2011.01729.x}}</ref> |
|||
The transition from early trophozoite to trophozoite to schizont correlates with the ordered induction of genes related to transcription/translation machinery, metabolic synthesis, energy metabolism, DNA replication, protein degradation, plastid functions, merozoite invasion, and motility. |
|||
The role of the apicoplast in the blood stages has been clarified.<ref name=Yeh2011>{{cite journal |author=Yeh E, Derisi JL |year=2011 |title=Chemical rescue of malaria parasites lacking an apicoplast defines organelle function in blood-stage ''Plasmodium falciparum'' |journal=PLoS Biol |volume=9 |issue=8 |page=e1001138 |doi=10.1371/journal.pbio.1001138 |editor1-last=Striepen |editor1-first=Boris}}</ref> Inhibition of [[isoprenoid]] precursor biosynthesis with the antibiotic [[fosmidomycin]] (an inhibitor of the enzyme [[DOXP reductoisomerase]]) causes delayed death in this parasite. This effect can be overcome with the addition of [[isopentenyl pyrophosphate]] (IPP) to the culture medium. Continued culture in the presence of this agent leads to the loss of the apicoplast genome and these mutants fail to process or localize organelle proteins. These auxotrophs can be grown indefinitely in asexual blood stage culture but are entirely dependent on exogenous IPP for survival. |
|||
Closely adjacent genes along the chromosome do not exhibit common transcription characteristics. Thus, genes are likely individually regulated along the parasite chromosome. |
|||
Iron-sulphur prosthetic groups are assembled in this organelle.<ref name=Kumar2011>{{cite journal |author=Kumar B, Chaubey S, Shah P, Tanveer A, Charan M, Siddiqi MI, Habib S |year=2011 |title=Interaction between sulphur mobilisation proteins SufB and SufC: Evidence for an iron-sulphur cluster biogenesis pathway in the apicoplast of ''Plasmodium falciparum'' |journal=Int J Parasitol |doi=10.1016/j.ijpara.2011.05.006 |volume=41 |issue=9 |pages=991–9 |pmid=21722645}}</ref> One component ([[SufB]]) is encoded in the apicoplast genome and a second ([[SufC]]) is encoded in the nucleus. SufB also exhibits ATPase activity. Other pathways that have been linked to this organelle include biosynthesis of isoprenoid precursors, fatty acids, heme and lipoic acid.<ref name=Seeber2010>Seeber F, Soldati-Favre D (2010) Metabolic pathways in the apicoplast of apicomplexa. Int Rev Cell Mol Biol 281: 161–228</ref> |
|||
Conversely, the apicoplast genome is polycistronic and most of its genes are coexpressed during the intraerythrocytic development cycle.<ref name="bozdech"/> |
|||
Iron-sulfur (Fe-S) cluster cluster synthesis pathways are found in the apicoplast.<ref name=Gisselberg2013>Gisselberg JE, Dellibovi-Ragheb TA, Matthews KA, Bosch G, Prigge ST (2013) The suf iron-sulfur cluster synthesis pathway is required for apicoplast maintenance in malaria parasites" ''PLoS Pathog'' 9(9) e1003655</ref> In the Suf pathway, SufS - a [[cysteine desulfurase]] - and its partner SufE are found exclusively in the apicoplast. In the Isc pathway IscS and its effector Isd11 were solely mitochondrial. Interruption of the Suf pathway results in a phenotpe that is dependent on external isopentenyl pyrophosphate. This phenotype is also associated with the loss of the apicoplast organelle and its organellar genome suggesting a role for this pathway in the maintenance of the apicoplast itself. |
|||
==Proteome== |
|||
There are 5,268 predicted proteins in ''[[Plasmodium falciparum]]'', and roughly 60% share little or no similarity to proteins in other organisms and thus are without functional assignment.<ref name="gardner"/> Of the predicted proteins, 31% contain at least one transmembrane domain, and 17.3% have a signal peptide or signal anchor.<ref name="gardner"/> |
|||
The [Fe-S] cluster protein NFUapi - a protein with a nitrogen fixation factor U (NifU)-like domain - is localised to the apicomplast.<ref name=Haussig2013>Haussig JM, Matuschewski K, Kooij TW (2013) Experimental genetics of ''Plasmodium berghei'' NFU in the apicoplast iron-sulfur cluster biogenesis pathway" ''PLoS One'' 8(6) e67269</ref> This protein may have a role in merosome formation and appears to be non essential for the blood stages. |
|||
It is estimated that 10.4% of the [[proteome]] is targeted to the [[apicoplast]].<ref name="gardner"/> |
|||
A gene [[''Plasmodium''-specific Apicoplast protein for Liver Merozoite]] formation (PALM) has been shown to be important for merozoite formation.<ref name=Haussig2011>Haussig JM, Matuschewski K, Kooij TW (2011) Inactivation of a ''Plasmodium'' apicoplast protein attenuates formation of liver merozoites. Mol Microbiol {{doi|10.1111/j.1365-2958.2011.07787.x}}</ref> Knock out mutants are unable to release merozoites into the blood from the liver stages. Mutants lacking this gene appear to be able to elicit at least temporary immunity. |
|||
It is estimated that 4.7% of the [[proteome]] is targeted to the mitochondria.<ref name="gardner"/> |
|||
Falcilysin a [[zinc]] [[metalloprotease]] is found in the apicoplast.<ref name=Ponpuak2006>{{cite journal |author=Ponpuak M, Klemba M, Park M, Gluzman IY, Lamppa GK, Goldberg DE |year=2006 |title=A role for falcilysin in transit peptide degradation in the ''Plasmodium falciparum'' apicoplast |journal=Mol Microbiol |volume=63 |issue=2 |pages=314–334 |doi=10.1111/j.1365-2958.2006.05443.x |pmid=17074076}}</ref> It is a member of the M16 protease group and has maximal activity at neutral pH. It appears to be an essential gene. Its function in this organelle is not quite clear but it appears to be involved in the degradation of transit peptides. |
|||
The parasite has different subsets of its proteome expressed during various stages of its developmental cycle.<ref name="florens">{{cite journal |
|||
| last = Florens |
|||
| first = |
|||
| authorlink = |
|||
| coauthors = |
|||
| title = A proteomic view of the ''Plasmodium falciparum'' life cycle |
|||
| journal = Nature |
|||
| volume = 419 |
|||
| issue = |
|||
| pages = 520–526 |
|||
| publisher = |
|||
| location = |
|||
| date = 3 October 2002 |
|||
| doi =10.1038/nature01107 |
|||
| id = |
|||
| accessdate = }}</ref> In one study, of the 2,415 proteins were identified in four stages(sporozoite, merozoite, trophozoite, gametocyte), representing 46% of the theoretical number of proteins.<ref name="florens"/> Only 6% of the proteins were found in all of the four stages. Of the proteins found, 51% were annotated as hypothetical proteins. |
|||
The enzyme [[thioredoxin peroxidase]] is found in the apicoplast, the mitochondrion and the cytosol.<ref name=Chaudhari2012>Chaudhari R, Narayan A, Patankar S (2012) A novel trafficking pathway in ''Plasmodium falciparum'' for the organellar localization of glutathione peroxidase-like thioredoxin peroxidase. FEBS J {{DOI|10.1111/j.1742-4658.2012.08746.x}}</ref> |
|||
[[Merozoites]] contained high levels of cell recognition and invasion proteins. [[Trophozoites]] contained proteins implicated in [[erythrocyte]] remodeling and [[hemoglobin]] digestion. Gametocytes contained high amounts of gametocyte-specific [[transcription factors]] and cell cycle/DNA processing proteins. The gametocytes had low levels of polymorphic surface antigens. Sporozoites contained large amounts of proteins related to invasion, as well as members of the ''var'' and ''rif'' families.<ref name="florens"/> |
|||
[[Autophagy]] is membrane-mediated degradation process that involves a series of proteins known as Atg proteins. [[Atg8]] is expressed during development and localises to the apicoplast.<ref name=Kitamura2012>Kitamura K, Kishi-Itakura C, Tsuboi T, Sato S, Kita K, Ohta N, Mizushima N (2012) Autophagy-related Atg8 localizes to the apicoplast of the human malaria parasite ''Plasmodium falciparum''" ''PLoS One'' 7(8) e42977.</ref> |
|||
Two C3 sugar phosphate transporter are present in the membrane of the apicoplast of ''[[Plasmodium berghei]]'' - [[triose phosphate]] transporter and [[phosphoenolpyruvate]] transporter.<ref name=Banerjee2012>Banerjee T, Jaijyan DK, Surolia N, Singh AP, Surolia A (2012) Apicoplast triose phosphate transporter (TPT) gene knockout is lethal for ''Plasmodium''. Mol Biochem Parasitol pii: S0166-6851(12)00241-1. {{DOI|10.1016/j.molbiopara.2012.09.008}}</ref> Knock out mutants of the triose phosphate transporter fail to survive. Phosphoenolpyruvate transporter knock out mutants survive in the blood stages but suffer defects during the liver stages development. |
|||
A pyruvate dehydrogenase complex with four subunits (E1alpha, E1beta, E2, E3) is present in the apicoplast.<ref name=Foth2005>Foth BJ, Stimmler LM, Handman E, Crabb BS, Hodder AN, McFadden GI (2005) The malaria parasite ''Plasmodium falciparum'' has only one pyruvate dehydrogenase complex, which is located in the apicoplast" ''Mol Microbiol'' 55(1) 39-53</ref> Unlike plants there is only a single copy of these genes in the genome. pyruvate dehydrogenase deficient parasites have no apparent blood stage growth defect, they are unable to progress beyond the oocyst phase of the parasite's mosquito stage.<ref name=Cobbold2013>Cobbold SA, Vaughan AM, Lewis IA, Painter HJ, Camargo N, Perlman DH, Fishbaugher M, Healer J, Cowman AF, Kappe SH, Llinás M (2013) Kinetic flux profiling elucidates two independent acetyl-CoA biosynthetic pathways in ''Plasmodium falciparum''. J Biol Chem </ref> |
|||
An ATP dependent caseinolytic protease (ClpP) is present in the apicomplast. Its function is currently unknown.<ref name=El_Bakkouri2012>El Bakkouri M, Rathore S, Calmettes C, Wernimont AK, Liu K, Sinha D, Asad M, Jung P, Hui R, Mohmmed A, Houry WA (2012) Structural insights into the inactive subunit of the apicoplast-localized caseinolytic protease complex of ''Plasmodium falciparum''. J Biol Chem</ref> |
|||
The lipid composition of the apicoplast has been analysed.<ref name="Botté2013">Botté CY, Yamaryo-Botté Y, Rupasinghe TW, Mullin KA, Macrae JI, Spurck TP, Kalanon M, Shears MJ, Coppel RL, Crellin PK, Maréchal E, McConville MJ, McFadden GI (2013) Atypical lipid composition in the purified relict plastid (apicoplast) of malaria parasites. Proc Natl Acad Sci USA</ref> some apicoplast lipids are generated de novo by the organelle itself. [[Phosphatidylinositol]] and other [[phospholipid]]s in the organelle are enriched in saturated [[fatty acid]]s. Lipids atypical for plastids - [[sphingomyelin]]s, [[ceramide]]s and [[cholesterol]] - are present. [[Galactoglycerolipid]]s, dominant in plant and algal plastids, are not present suggesting that these glycolipids are a hallmark of photosynthetic plastids and were lost when these organisms assumed a parasitic lifestyle. |
|||
The endoplasmic reticulum associated degradation ([[Endoplasmic-reticulum-associated protein degradation|ERAD]]) machinery is a quality control mechanism that retro-translocates misfolded secretory proteins across the endoplasmic reticulum membrane.<ref name=Smith2011>Smith MH, Ploegh HL, Weissman JS (2011) Road to ruin: targeting proteins for degradation in the endoplasmic reticulum" ''Science'' 334: 1086–1090. {{DOI|10.1126/science.1209235}}</ref> Several components of this system including the membrane protein [[Der1]], the AAA ATPase [[Cdc48]] and its cofactor [[Ufd1]] appear to be present in the apicoplast and are involved in transmembrane transport. |
|||
[[ClpB]] is a molecular chaperone and a member of the AAA+ superfamily of ATPases. There are 2 isoforms of ClpB (PfClpB1 and PfClpB2) found in the apicoplast.<ref name=Ngansop2013>Ngansop F, Li H, Zolkiewska A, Zolkiewski M (2013) Biochemical characterization of the apicoplast-targeted AAA+ ATPase ClpB from ''Plasmodium falciparum''. Biochem Biophys Res Commun pii: S0006-291X(13)01411-3. {{DOI|10.1016/j.bbrc.2013.08.064}}</ref> PfClpB1 contains all characteristic AAA+sequence motifs but there is a 52-residue long non-conserved insert middle domain. ATP induces self association of PfClpB1 into hexamers like in most AAA+ ATPases. It catalyzes the hydrolysis of ATP and its ATPase activity is activated in the presence of [[casein]] and polylysine. |
|||
A glutamyl-tRNA synthetase (GluRS) is present in the apicoplast.<ref name=Mailu2013>Mailu BM, Ramasamy G, Mudeppa DG, Li L, Lindner SE, Peterson MJ, Derocher AE, Kappe SH, Rathod PK, Gardner MJ (2013) A non-discriminating glutamyl-tRNA synthetase in the ''Plasmodium'' apicoplast: The first enzyme in an indirect aminoacylation pathway. J Biol Chem</ref> It is it is non-discriminating glutamylating both apicoplast tRNA-Glu and tRNA-Gln. It appears to be an essential enzyme. |
|||
An elongation factor G which is involved in translation is present in the apicoplast.<ref name=Gupta2013>Gupta A, Mir SS, Saqib U, Biswas S, Vaishya S, Srivastava K, Siddiqi MI, Habib S (2013) The effect of fusidic acid on ''Plasmodium falciparum'' elongation factor G (EF-G). Mol Biochem Parasitol. 2013 Nov 6. pii: S0166-6851(13)00151-5. doi: 10.1016/j.molbiopara.2013.10.003 </ref> It has GTPase activity that can be inhibited by [[fusidic acid]]. |
|||
===Mitochondrion=== |
|||
''Plasmodium'' lacks mitochondrial [[pyruvate dehydrogenase]]<ref>{{cite journal |author=McMillan PJ, Stimmler LM, Foth BJ, McFadden GI, Müller S |title=The human malaria parasite ''Plasmodium falciparum'' possesses two distinct dihydrolipoamide dehydrogenases |journal=[[Molecular Microbiology]] |volume=55 |issue=1 |pages=27–38 |year=2005 |month=January |pmid=15612914 |doi=10.1111/j.1365-2958.2004.04398.x |url=http://onlinelibrary.wiley.com/resolve/openurl?genre=article&sid=nlm:pubmed&issn=0950-382X&date=2005&volume=55&issue=1&spage=27}}</ref><ref>{{cite journal |author=Foth BJ, Stimmler LM, Handman E, Crabb BS, Hodder AN, McFadden GI |title=The malaria parasite Plasmodium falciparum has only one pyruvate dehydrogenase complex, which is located in the apicoplast |journal=[[Molecular Microbiology]] |volume=55 |issue=1 |pages=39–53 |year=2005 |month=January |pmid=15612915 |doi=10.1111/j.1365-2958.2004.04407.x |url=http://onlinelibrary.wiley.com/resolve/openurl?genre=article&sid=nlm:pubmed&issn=0950-382X&date=2005&volume=55&issue=1&spage=39}}</ref> and the hydrogen ion translocating NADH [[dehydrogenase]] (Complex I, NDH1). The [[mitochondrion]] contains a minimal DNA genome (~6 kilobases) and carries out oxidative phosphorylation in the insect vector stages by using [[2-oxoglutarate]] as an alternative means of entry into the [[tricarboxylic acid cycle]] and a single-subunit flavoprotein as an alternative NADH dehydrogenase (NDH2). In the blood stages mitochondrial enzymes are down regulated and parasite energy metabolism relies mainly on glycolysis. The enzyme [[malate quinone oxidoreductase]] was acquired from an epsilon [[proteobacteria]] via lateral gene transfer. This transfer occurred in an ancestor of the Apicomplexa. |
|||
The [[ATP synthase]] is localised to the mitochondrion, is assembled as a large dimeric complex and appears to be essential for in the blood stages of the life cycle.<ref>{{cite journal |author=Balabaskaran Nina P, Morrisey JM, Ganesan SM, Ke H, Pershing AM, Mather MW, Vaidya AB |year=2011 |title=ATP synthase complex of ''Plasmodium falciparum'': dimeric assembly in mitochondrial membranes and resistance to genetic disruption |journal=J Biol Chem}}</ref> Its function in these stages is not yet clear. |
|||
The [[GFER|sulfhydryl:cytochrome c oxidoreductase Erv1/ALR/GFER/HSS]] (Essential for Respiration and Vegatative growth/Augmenter of Liver Regeneration/Growth Factor Erv1-like/Hepatic regenerative Stimulation Substance/hepatopoietin) is an essential sulfhydryl oxidase for required oxidative protein import into the mitochondrial intermembrane space. It is one of several enzymes involved in electron transferase activity. It is encoded by all eukaryotes and cytoplasmic DNA viruses sequenced to date. The enzyme from ''P. falciparum'' differs significantly from that found in yeast and humans with altered cysteine motifs and intermolecular disulfide bonds.<ref name=Eckers2012>Eckers E, Petrungaro C, Gross D, Riemer J, Hell K, Deponte M (2012) Divergent molecular evolution of the mitochondrial sulfhydryl:cytochrome c oxidoreductase Erv in opisthokonts and parasitic protists. J Biol Chem</ref> Despite successful cloning and expression in yeast, the parasite enzyme fails to function in yeast. A second related enzyme - Mia40 - does not appear to be present in ''P. falciparum''. |
|||
Deletion of the gene in the rodent parasite ''[[Plasmodium berghei]]'' for the [[flavoprotein]] subunit of [[succinate dehydrogenase]] - part of the complex II - showed impairment of ookinete function and oocyst formation.<ref name=Hino2012>Hino A, Hirai M, Tanaka TQ, Watanabe YI, Matsuoka H, Kita K (2012) Critical roles of the mitochondrial complex II in oocyst formation of rodent malaria parasite ''Plasmodium berghei''. J Biochem</ref> |
|||
The gene for the flavoprotein subunit of [[succinate dehydrogenase]] can be disrupted in the parasite.<ref name=Tanaka2012>Tanaka TQ, Hirai M, Watanabe YI, Kita K (2012) Towards understanding the role of mitochondrial complex II in the intraerythrocytic stages of ''Plasmodium falciparum'': Gene targeting of the Fp subunit. Parasitol Int</ref> Its disruption causes growth retardation of the intraerythrocytic forms. It appears that complex II functions as a quinol-fumarate reductase to form [[succinate]] from [[fumarate]] in the intraerythrocytic parasite. |
|||
The [[dicarboxylate-tricarboxylate carrier]] homolog has been cloned from ''P. falciparum''.<ref name=Nozawa2011>Nozawa A, Fujimoto R, Matsuoka H, Tsuboi T, Tozawa Y (2011) Biochem Biophys Res Commun</ref> This protein may mediate the [[Alpha-Ketoglutaric acid|oxoglutarate]]-[[malate]] exchange across the inner mitochondrial membrane required for the branched pathway of tricarboxylic acid metabolism. |
|||
The ClpQ protease and ClpY ATPase have been cloned.<ref>{{cite journal |author=Rathore S, Jain S, Sinha D, Gupta M, Asad M, Srivastava A, Narayanan MS, Ramasamy G, Chauhan VS ''et al.'' |year=2011 |title=Disruption of a mitochondrial protease machinery in ''Plasmodium falciparum'' is an intrinsic signal for parasite cell death |journal=Cell Death Dis |volume=2 |issue=11 |page=e231 |doi=10.1038/cddis.2011.118}}</ref> ClpQY function disruption caused hindrance in the parasite growth and maturation of asexual stages of parasites. Features of apoptosis like cell death are also found. |
|||
The mitochondrial pathway of protein lipoylation relies on scavenging from the host and can be inhibited with the lipoic acid analog 8-bromo-octanoic acid. Use of this agent inhibits growth and significantly reduces [[merosome]] formation. Schizogony is the phase most affected by this inhibition.<ref name=Deschermeier2011>Deschermeier C, Hecht LS, Bach F, Rützel K, Stanway RR, Nagel A, Seeber F, Heussler VT (2011) Mitochondrial lipoic acid scavenging is essential for Plasmodium berghei liver stage development. Cell Microbiol {{doi|10.1111/j.1462-5822.2011.01729.x}}</ref> |
|||
[[Atovaquone]], a 2-hydroxynaphthoquinone, is a competitive [[Enzyme inhibitor|inhibitor]] of the [[quinol]] oxidation site of the mitochondrial cytochrome bc1 complex and is used as an antimalaria agent.<ref name=Fisher2012>{{cite journal |author=Fisher N, Abd Majid R, Antoine T, Al-Helal M, Warman AJ, Johnson DJ, Lawrenson AS, Ranson H, O'Neill PM, Ward SA, Biagini GA |year=2012 |title=Cytochrome b mutation Y268S conferring the atovaquone resistance phenotype in the malaria parasite results in reduced parasite bc1 catalytic turnover and protein expression |journal=J Biol Chem |doi=10.1074/jbc.M111.324319 |volume=287 |issue=13 |pages=9731–41 |pmid=22282497 |pmc=3322985}}</ref> Inhibition of this enzyme leads to the collapse of the mitochondrial membrane potential and disruption of pyrimidine biosynthesis. These effects are lethal to the parasite. |
|||
The mitochondrial RNA polymerase appears to be an essential gene for the erythrocytic stages.<ref name=Ke2012>Ke H, Morrisey J, Ganesan SM, Mather MW, Vaidya AB (2012) Mitochondrial RNA polymerase is an essential enzyme in erythrocytic stages of Plasmodium falciparum. Mol Biochem Parasitol</ref> |
|||
Over half the genome of the mitochondrion encodes the genes for three classic mitochondrial proteins: [[cytochrome oxidase]] subunits I and III and [[apocytochrome b]].<ref name=Feagin2012>Feagin JE, Harrell MI, Lee JC, Coe KJ, Sands BH, Cannone JJ, Tami G, Schnare MN, Gutell RR (2012) The fragmented mitochondrial ribosomal RNAs of ''Plasmodium falciparum''" ''PLoS One'' 7(6) e38320.</ref> The remainder encodes 34 RNA genes of which 27 have been assigned to ribosomal RNA (12 to the small subunit and 15 to the large subunit). These genes are fragmented and are encoded on both strands. |
|||
The mitochondrial [[thioredoxin peroxidase]]-2 does not appear to be essential.<ref name=Masuda-Suganuma2012>Masuda-Suganuma H, Usui M, Fukumoto S, Inoue N, Kawazu SI (2012) Mitochondrial peroxidase TPx-2 is not essential in the blood and insect stages of ''Plasmodium berghei''. Parasit Vectors 5(1) 252</ref> |
|||
The ATP dependent ClpQY system is a prokaryotic proteasome like multisubunit machinery localized in the mitochondrion. It has two components - a ClpQ threonine protease and ClpY ATPase. The ClpQ threonine protease appears to be an essential gene.<ref name=Jain2013>Jain S, Rathore S, Asad M, Hossain ME, Sinha D, Datta G, Mohmmed A (2013) The prokaryotic ClpQ protease plays a key role in growth and development of mitochondria in ''Plasmodium falciparum''. Cell Microbiol {{DOI|10.1111/cmi.12142}}</ref> |
|||
The bc 1 complex of the mitochondrial respiratory chain is essential for the parasite's life cycle. The drug [[atovaquone]] inhibits its activity. Atovaquone binds to the Q<sub>0</sub> site on this complex.<ref name="Vallières2013">Vallières C, Fisher N, Meunier B (2013) Reconstructing the Qo Site of ''Plasmodium falciparum'' bc 1 complex in the yeast enzyme" ''PLoS One'' 8(8) e71726. {{DOI|10.1371/journal.pone.0071726}}</ref> |
|||
An AAA+/[[FtsH protease]] homolog (PfFtsH1) that exhibits ATP- and Zn<sup>2+</sup>-dependent protease activity is present in the inner mitochondrial membrane.<ref name=Tanveer2013>Tanveer A, Allen SM, Jackson KE, Charan M, Ralph SA, Habib S (2013) An FtsH protease is recruited to the mitochondrion of ''Plasmodium falciparum''" ''PLoS One'' 8(9) e74408. {{DOI|10.1371/journal.pone.0074408}}</ref> It seems likely that this is a regulatory protein for organelle biogenesis. |
|||
An [[elongation factor]] G involved in protein translation is present in the mitochondrion.<ref name=Gupta2013>Gupta A, Mir SS, Saqib U, Biswas S, Vaishya S, Srivastava K, Siddiqi MI, Habib S (2013) The effect of fusidic acid on ''Plasmodium falciparum'' elongation factor G (EF-G). Mol Biochem Parasitol. 2013 Nov 6. pii: S0166-6851(13)00151-5. doi: 10.1016/j.molbiopara.2013.10.003 </ref> It has GTPase activity that can be inhibited by [[fusidic acid]]. |
|||
===Digestive vacuole=== |
|||
During growth of the parasite and as part of its digestion of the erythrocyte's haemoglobin, fusion of digestive vesicles occurs and gives rise to a large digestive vacuole.<ref name=Wunderlich2012>Wunderlich J, Rohrbach P, Dalton JP (2012) The malaria digestive vacuole. Front Biosci (Schol Ed) 4:1424-1448</ref> This vacuole the interior of which is maintained at a low pH (pH 4.5 - 5.5), processes 60-80% of the ingested hemoglobin and provides a pool of amino acids that is crucial for parasite growth and development. The membrane contains ion pumps and transporters that maintain its low pH. During haemoglobin digestion the [[heme]] is released from hemoglobin. Haem is toxic to the parasite and is detoxified by biocrystallization to hemozoin within the vacuole. Quinoline drugs, including chloroquine, act by binding to heme and thus prevent its sequestration into hemozoin. |
|||
It has been shown that micromolar concentrations of [[chloroquine]] partially permeabilized the parasite's digestive vacuole membrane and that this event appears to precede mitochondrial dysfunction.<ref name=Ch>{{cite journal |author=Ch'ng JH, Liew K, Goh AS, Sidhartha E, Tan KS |year=2011 |title=Drug-induced permeabilization of parasite's digestive vacuole is a key trigger of programmed cell death in ''Plasmodium falciparum'' |journal=Cell Death Dis |volume=2 |issue=10 |page=e216 |doi=10.1038/cddis.2011.97}}</ref> |
|||
[[Quinine]] has been shown localise to a non acidic compartment within the digestive vacuole.<ref name="Bohórquez2012">Bohórquez EB, Chua M, Meshnick SR (2012) Quinine localizes to a non-acidic compartment within the food vacuole of the malaria parasite ''Plasmodium falciparum''. Malar J</ref> It may colocate with haemozoin. It's localisation within the parasite is not altered by the presence or absence of a functional multidrug resistance gene. |
|||
The digestive vacuole is able to activate both the alternative [[Complement system|complement]] and the intrinsic clotting pathway.<ref name=Dasari2012>Dasari P, Heber SD, Beisele M, Torzewski M, Reifenberg K, Orning C, Fries A, Zapf AL, Baumeister S, Lingelbach K, Udomsangpetch R, Bhakdi SC, Reiss K, Bhakdi S (2012) Digestive vacuole of ''Plasmodium falciparum'' released during erythrocyte rupture dually activates complement and coagulation. Blood</ref> The digestive vacuole membrane has the capacity to assemble [[prothrombinase]], a key enzyme of the intrinsic clotting pathway.<ref name=Dasari2012>Dasari P, Bhakdi S (2012) Pathogenesis of malaria revisited. Med Microbiol Immunol</ref> The capacity of this membrane to activate both complement and coagulation can be suppressed by low molecular weight [[dextran sulfate]]. Phagocytosis of these membranes drives the [[Granulocyte|polymorphonucleocyte]]s into a state of functional exhaustion. |
|||
Two multi-spanning digestive vacuole membrane proteins are known: the multidrug resistance protein 1 and Chloroquine Resistance Transporter (CRT).<ref name=Ehlgen2012>{{cite journal |last1=Ehlgen |first1=F |last2=Pham |first2=JS |last3=De Koning-Ward |first3=T |last4=Cowman |first4=AF |last5=Ralph |first5=SA |year=2012 |title=Investigation of the ''Plasmodium falciparum'' food vacuole through inducible expression of the chloroquine resistance transporter (PfCRT) |journal=PLoS ONE |volume=7 |issue=6 |page=e38781 |doi=10.1371/journal.pone.0038781 |editor1-last=Snounou |editor1-first=Georges}}</ref> The CRT protein moves from the endoplasmic reticulum to the Golgi apparatus before becoming associated with the digestive vacuole. The digestive vacuole forms in the ring stages of the parasites life cycle. Chloroquine sensitivity is not influenced by the absence of CRT from the digestive vacuole bringing into question its relationship (if any) to chloroquine resistance.<ref name="Ehlgen2012"/> Mutations in the CRT gene have been associated with sensitivity and resistance to quinolines.<ref>Griffin CE, Hoke JM, Samarakoon U, Duan J, Mu J, Ferdig MT, Warhurst DC, Cooper RA (2012) Mutation in the ''Plasmodium falciparum'' CRT protein determines the stereospecific activity of the antimalarial Cinchona alkaloids. Antimicrob Agents Chemother</ref> In particular a [[lysine]] to [[isoleucine]] at [[codon]] 76 ([[Adenosine]] -> [[Thymine]] at base 227) mutation and a [[valine]] to [[phenylalanine]] ([[Guanine]] -> Thymine at base 1108) mutation have been associated with changes in drug sensitivity. The mutant CRT protein increases the transport of glutathione into the digestive vacuole. The role - if any - that this has on chloroquine resistance is presently unknown. |
|||
The membrane of the digestive vacuole is four [[nanometer]]s in thickness with patches that may be up to 12 nanometers in thickness.<ref name=Kapishnikov2012>Kapishnikov S, Weiner A, Shimoni E, Guttmann P, Schneider G, Dahan-Pasternak N, Dzikowski R, Leiserowitz L, Elbaum M (2012) Oriented nucleation of hemozoin at the digestive vacuole membrane in ''Plasmodium falciparum''. Proc Natl Acad Sci USA</ref> |
|||
The digestion of haemoglobin produces large quantities of [[ferriprotoporphyrin]] IX which it unable to digest and is potentially toxic to the parasite. To avoid the toxicity the ferriprotoporphyrin is converted to haemozoin. Chloroquine inhibits this process. The mechanisms behind this process are still unclear. ''In vitro'' conversion of ferriprotoporphyrin to haemazoin is enhanced at a temperature of 41C when compared to its conversion at 37C.<ref name=Orjih2012>Orjih AU, Mathew TC, Cherian PT (2012) Erythrocyte membranes convert monomeric ferriprotoporphyrin IX to β-hematin in acidic environment at malarial fever temperature. Exp Biol Med (Maywood)</ref> It is possible that the rise in temperature that occurs in malaria may be part of a strategy to enhance this reaction at the later stages of growth when the ferriprotoporphyrin concentration is likely to be high. |
|||
Inhibition of the falcipains involved in the digestion of haemoglobin results in enlargement of the digestive vacuole.<ref name=Prasad2013>Prasad R, Atul, Kolla VK, Legac J, Singhal N, Navale R, Rosenthal PJ, Sijwali PS (2013) Blocking ''Plasmodium falciparum'' development via dual inhibition of hemoglobin degradation and the ubiquitin proteasome system by MG132" ''PLoS One'' 8(9) e73530. {{DOI|10.1371/journal.pone.0073530}}</ref> |
|||
===Nucleus=== |
|||
Unlike most other eukaryotes the chromosomes never become condensed even during mitosis and remain difficult to visualise by llight microscopy. |
|||
Studies with antibodies to the nuclear pore protein subunit PfNup116 show that the nuclear pores are highly polarized during the ring and schizont stages whereas in trophozoite stage the nuclear pores redistributed over the entire nuclear surface.<ref name=Guizetti2013>Guizetti J, Martins RM, Guadagnini S, Claes A, Scherf A (2013) Nuclear pores and perinuclear expression sites of ''var'' and rDNA genes correspond to physically distinct regions in ''Plasmodium falciparum''. [[Eukaryotic Cell]]</ref> |
|||
PfSec13 is a [[nucleoporin]].<ref name=Dahan-Pasternak2013>Dahan-Pasternak N, Nasereddin A, Kolevzon N, Pe'er M, Wong W, Shinder V, Turnbull L, Whitchurch CB, Elbaum M, Gilberger TW, Yavin E, Baum J, Dzikowski R (2013) PfSec13 is an unusual chromatin associated nucleoporin of ''Plasmodium falciparum'', which is essential for parasite proliferation in human erythrocytes. J Cell Sci</ref> It appears to be a fusion between Sec13 and Nup145C and it associates with the [[nuclear pore complex]]es and [[microtubule]]s. It appears to be an essential gene. |
|||
===Nucleolus=== |
|||
A hat-like structure polarized towards one side of the nucleus that stains with nucleolar markers has been described<ref name=Figueiredo2005>{{cite journal |author=Figueiredo LM, Rocha EP, Mancio-Silva L, Prevost C, Hernandez-Verdun D, Scherf A |year=2005 |title=The unusually large ''Plasmodium'' telomerase reverse-transcriptase localizes in a discrete compartment associated with the nucleolus |journal=Nucleic Acids Res |volume=33 |issue=3 |pages=1111–1122 |doi=10.1093/nar/gki260 |pmid=15722485 |pmc=549419}}</ref> It seems likely that this unusual structure is the [[nucleolus]]. |
|||
The histone deacetylase Sir2a is found in the nucleolus in addition to the telomeres.<ref name=Mancio-Silva2013>Mancio-Silva L, Lopez-Rubio JJ, Claes A, Scherf A (2013) Sir2a regulates rDNA transcription and multiplication rate in the human malaria parasite ''Plasmodium falciparum''. Nat Commun 4:1530. {{DOI|10.1038/ncomms2539}}</ref> It functions there to control the transcription of the ribosomal RNA levels. |
|||
The ribosomal RNA genes are found localised to this organelle.<ref name=Mancio-Silva2012>Mancio-Silva L, Zhang Q, Scheidig-Benatar C, Scherf A (2010) Clustering of dispersed ribosomal DNA and its role in gene regulation and chromosome-end associations in malaria parasites" ''Proc Natl Acad Sci USA'' 107(34) 15117-15122. {{DOI|10.1073/pnas.1001045107}}</ref> |
|||
===Endoplasmic reticulum=== |
|||
This forms a set of reticular structures adjacent to the nuclear regions during the trophozoite and schizont stages.<ref name=Chung2012>Chung DW, Ponts N, Prudhomme J, Rodrigues EM, Le Roch KG (2012) Characterization of the ubiquitylating components of the human malaria parasite's protein degradation pathway" ''PLoS One'' 7(8) e43477</ref> In the late schizont stage it forms globular structures surrounding each budding merozoite. |
|||
A Ca<sup>2+</sup> [[ATPase]] 6 has been associated with resistance to artemisinin resistance.<ref name=Huang2012>Huang F, Tang L, Yang H, Zhou S, Liu H, Li J, Guo S (2012) Molecular epidemiology of drug resistance markers of ''Plasmodium falciparum'' in Yunnan Province, China. Malar J 11:243</ref> The gene has a single copy in the genome.<ref name=Phompradit2012>Phompradit P, Wisedpanichkij R, Muhamad P, Chaijaroenkul W, Na-Bangchang K (2012) Molecular analysis of pfatp6 and pfmdr1 polymorphisms and their association with in vitro sensitivity in ''Plasmodium falciparum'' isolates from the Thai-Myanmar border. Acta Trop. 2011 Oct-Nov;120(1-2) 130-5. {{DOI|10.1016/j.actatropica.2011.07.003}}</ref> Reistance has been associated with four mutations: [[codon]] 263 [[Lysine]]->[[Glutamic acid]], codon 431 Glutamic acid->Lysine, codon 623 [[Alanine]]->Glutamic acid and codon 769 [[Serine]]->[[Asparagine]].<ref name=Tahar2009>Tahar R, Ringwald P, Basco LK (2009) Molecular epidemiology of malaria in Cameroon. XXVIII. In vitro activity of dihydroartemisinin against clinical isolates of ''Plasmodium falciparum'' and sequence analysis of the ''P. falciparum'' ATPase 6 gene" ''Am J Trop Med Hyg'' 81(1) 13-18</ref> This gene is a SERCA pump and appears to be essential in the asexual stages.<ref name=Pulcini2013>Pulcini S, Staines HM, Pittman JK, Slavic K, Doerig C, Halbert J, Tewari R, Shah F, Avery MA, Haynes RK, Krishna S (2013) Expression in yeast links field polymorphisms in PfATP6 to in vitro artemisinin resistance and identifies new inhibitor classes. J Infect Dis</ref> |
|||
Many proteins in the genome carry a host targeting signal.<ref name=Bhattacharjee2012>{{cite journal |author=Bhattacharjee S, Stahelin RV, Speicher KD, Speicher DW, Haldar K |year=2012 |title=Endoplasmic reticulum PI(3)P lipid binding targets malaria proteins to the Host Cell |journal=Cell |volume=148 |issue=1–2 |pages=201–212 |doi=10.1016/j.cell.2011.10.051 |pmid=22265412 |pmc=3268671}}</ref> This signal sequence is recognised by [[phosphatidylinositol-3-phosphate]] in the [[endoplasmic reticulum]]. |
|||
===Ribosomes=== |
|||
Unlike other eukaryotes studied to date ''[[Plasmodium]]'' species have two or three distinct SSU rRNA (18S rRNA) molecules encoded within the genome.<ref name="Nishimoto2008">{{cite journal |last1=Nishimoto |first1=Y |last2=Arisue |first2=N |last3=Kawai |first3=S |last4=Escalante |first4=AA |last5=Horii |first5=T |last6=Tanabe |first6=K |last7=Hashimoto |first7=T. |author-separator=, |year=2008 |title=Evolution and phylogeny of the heterogeneous cytosolic SSU rRNA genes in the genus ''Plasmodium'' |journal=Mol Phylogenet Evol. |volume=47 |issue=1 |pages=45–53 |doi=10.1016/j.ympev.2008.01.031 |pmid=18334303}}</ref> These have been divided into types A, S and O. Type A is expressed in the asexual stages; type S in the sexual and type O only in the oocyte. Type O is only known to occur in ''[[Plasmodium vivax]]'' at present. The reason for this gene duplication is not known but presumably reflects an adaption to the different environments the parasite lives within. |
|||
The Asian simian ''Plasmodium'' species - ''[[Plasmodium coatneyi]]'', ''[[Plasmodium cynomolgi]]'', ''[[Plasmodium fragile]]'', ''[[Plasmodium inui]]'', ''[[Plasmodium fieldi]]'', ''[[Plasmodium hylobati]]'' and ''[[Plasmodium simiovale]]'' - have a single single S-type-like gene and several A-type-like genes. Phylogenetic analyses has shown that gene duplication events giving rise to A- and S-type-like sequences took place independently at least three times in the ''Plasmodium'' evolution. |
|||
The phosphoprotein [[RPLP0|P0]] occurs as a complex with two other small acidic ribosomal proteins ([[RPLP1|P1]] and [[RPLP2|P2]]).<ref name=Das2012>Das S, Basu H, Korde R, Tewari R, Sharma S (2012) Arrest of nuclear division in ''Plasmodium'' through blockage of erythrocyte surface exposed ribosomal protein P2" ''PLoS Pathog'' 8(8) e1002858</ref> A pentameric complex [(P1–P2) P0 (P1–P2)] form the stalk of the large ribosomal subunit, which seems to play a role in the [[GTPase]] elongation centre of the ribosome. |
|||
The P2 protein is exported to the infected erythrocyte surface at 30 hrs post merozoite invasion, concomitant with extensive oligomerization. It is largely largely composed of alpha helical and random coil domains.<ref name=Das2012>Das S, Rajagopal S, Sivakami S, Sharma S (2012) Erythrocytic stage dependent regulation of oligomerization of ''Plasmodium'' ribosomal protein P2. J Biol Chem</ref> |
|||
===Acidocalcisomes=== |
|||
Acidocalcisomes are acidic calcium stores and are present in many organisms including [[bacteria]], ''Plasmodium'' and humans.<ref name=Rohloff2011>Rohloff P, Miranda K, Rodrigues JC, Fang J, Galizzi M, Plattner H, Hentschel J, Moreno SN (2011) Calcium uptake and proton transport by acidocalcisomes of ''Toxoplasma gondii''" ''PLoS One'' 6(4) e18390. {{DOI|10.1371/journal.pone.0018390}}</ref> The organelles possess an acidic matrix that contains several cations bound to phosphates. These are mainly present in the form of short and long polyphosphate chains. The matrix is acidified through the action of proton pumps such as a vacuolar proton ATPase and a vacuolar proton pyrophosphatase. Calcium uptake occurs through a Ca<sup>2+</sup>/H<sup>+</sup> countertransporting ATPase located in the membrane of the organelle. |
|||
===Motility=== |
|||
Thrombospondin Related Anonymous Protein (TRAP) is a type I transmembrane proteins which has several extracellular adhesive domains and a cytoplasmic domain that recruits the glycolytic enzyme [[aldolase]]. Normally only small amount of TRAP found on the sporozoite surface. TRAP is involved in cell motility.<ref name=Hellmann2011>{{cite journal |author=Hellmann JK |year=2011 |title=Environmental constraints guide migration of malaria parasites during transmission |journal=PLoS Pathog |volume=7 |issue=6 |page=e1002080 |author-separator=, |author2=Münter S |author3=Kudryashev M |author4=Schulz S |author5=Heiss K |author6=Müller AK |author7=Matuschewski K |author8=Spatz JP |author9=Schwarz US |display-authors=9 |doi=10.1371/journal.ppat.1002080 |editor1-last=Mota |editor1-first=Maria M |first10=Friedrich}}</ref> |
|||
Its tandem von Willebrand factor A and thrombospondin type I repeat domains connect through the [[proline]] rich stalk, transmembrane and cytoplasmic domains to the parasite's actin dependent motility apparatus.<ref name=Song2012>Song G, Koksal AC, Lu C, Springer TA (2012) Shape change in the receptor for gliding motility in ''Plasmodium'' sporozoites. Proc Natl Acad Sci USA</ref> Binding is dependent on the presence of a metal ion. The protein is capable of considerable conformational changes. There is also a potential heparan sulphate binding site in the von Willebrand factor A domain.<ref name=Pihlajamaa2013>Pihlajamaa T, Kajander T, Knuuti J, Horkka K, Sharma A, Permi P (2013) Structure of ''Plasmodium falciparum'' thrombospondin-related anonymous protein (TRAP) A domain highlights distinct features in apicomplexan von Willebrand Factor A homologues. Biochem J</ref> |
|||
The cytoplasmic domain binds to F-[[actin]] which connects to [[myosin]] A. Within the transmembrane domain it has a canonical rhomboid cleavage site ([[Alanine|Ala]]-[[Glycine|Gly]]-Gly-[[Isoleucine|Ile]]-Ile-Gly-Gly). [[Rhomboid protease]]s are a family of serine proteases that require helical instability in the transmembrane domain and have specific residue requirements in their P1, P4 and P2′ positions. These proteases are responsible for intramembraneous cleavage. |
|||
TRAP binds to receptors on the host and is translocated posteriorly by the actomyosin motor. It is then normally cleaved by a calcium independent serine protease. Removal of the cytoplasmic domain abolish the motility of the parasite. Mutations in the rhomboid cleavage site are defective in TRAP shedding and display slow, staccato motility and reduced infectivity.<ref name=Ejigiri2012>Ejigiri I, Ragheb DR, Pino P, Coppi A, Bennett BL, Soldati-Favre D, Sinnis P (2012) Shedding of TRAP by a rhomboid protease from the malaria sporozoite surface is essential for gliding motility and sporozoite infectivity" ''PLoS Pathog'' 8(7) e1002725</ref> The reduction in infectivity is particularly marked if the sporozoites are inoculated intradermally rather than intravascularly. Prevention of cleavage of the TRAP protein entirely renders the sporozoites uninfectious and immobile. The rhomboid protease normally involved in TRAP cleavage appears to be the ROM4 protease. This protease is found across the entire sporozoite surface suggesting it has functions in addition to TRAP cleavage. |
|||
The circumsporozoite- and thrombospondin-related adhesive protein (CTRP) is a modular multidomain protein containing six tandem [[von Willebrand factor]] A like domains and seven tandem [[thrombospondin]] type I repeat-like domains.<ref name=Ramakrishnan2011>{{cite journal |author=Ramakrishnan C, Dessens JT, Armson R, Pinto SB, Talman AM, Blagborough AM, Sinden RE |year=2011 |title=Vital functions of the malarial ookinete protein, CTRP, reside in the A domains |journal=Int J Parasitol |doi=10.1016/j.ijpara.2011.05.007 |volume=41 |issue=10 |pages=1029–39 |pmid=21729699}}</ref> The A domains of CTRP are critical for ookinete gliding motility and oocyst formation. The thrombospondin domains are fully redundant. |
|||
The cell-traversal protein for ookinetes and sporozoites (CelTOS) is a protein involved in the invasion of both vertebrate and insect host cells.<ref name=Bergmann-Leitner2011>{{cite journal |author=Bergmann-Leitner ES, Legler PM, Savranskaya T, Ockenhouse CF, Angov E |year=2011 |title=Cellular and humoral immune effector mechanisms required for sterile protection against sporozoite challenge induced with the novel malaria vaccine candidate CelTOS |journal=Vaccine |doi=10.1016/j.vaccine.2011.06.053 |volume=29 |issue=35 |pages=5940–9 |pmid=21722682}}</ref> |
|||
The C-terminal tail of myosin A (MyoA) and its light chain, myosin A tail domain interacting protein (MTIP) are essential parts of the gliding motility apperatus.<ref name=Douse2012>Douse CH, Green JL, Salgado PS, Simpson PJ, Thomas JC, Langsley G, Holder AA, Tate EW, Cota E (2012) Regulation of the ''Plasmodium'' motor complex: phosphorylation of Myosin A Tail Interacting Protein (MTIP) loosens its grip on MyoA. J Biol Chem</ref> |
|||
[[Dynein]] light chain 8 is present in ''P. falciparum'' as a homodimer.<ref name=Qureshi2012>Qureshi BM, Hofmann NE, Arroyo-Olarte RD, Nickl B, Hoehne W, Jungblut PR, Lucius R, Scheerer P, Gupta N (2012) Dynein light chain 8a of ''Toxoplasma gondii'', a unique conoid-localized β-strand-swapped homodimer, is required for an efficient parasite growth. FASEB J</ref> The dimer is formed by the interaction of the β<sub>0</sub> chains on one molecule with the β<sub>2</sub> chains of the second. |
|||
A [[kinesin]] with the ability to depolymerise [[microtubule]]s has been cloned.<ref name=Shipley2004>Shipley K, Hekmat-Nejad M, Turner J, Moores C, Anderson R, Milligan R, Sakowicz R, Fletterick R () Structure of a kinesin microtubule depolymerization machine" ''EMBO J'' 23(7) 1422-1432</ref> |
|||
===Autophagy=== |
|||
A number of proteins involved in [[autophagy]] are known to be present in the genome. These include Atg8 and Atg3.<ref name=Hain2012>Hain AU, Weltzer RR, Hammond H, Jayabalasingham B, Dinglasan RR, Graham DR, Colquhoun DR, Coppens I, Bosch J (2012) Structural characterization and inhibition of the Plasmodium Atg8-Atg3 interaction. J Struct Biol pii: S1047-8477(12)00243-2. {{DOI|10.1016/j.jsb.2012.09.001}}</ref> The functions of these proteins in the parasite are still being elucidated. |
|||
Autophagosomes fuse with the endosomes before being routed to the digestive vacuole.<ref name=Tomlins2013>Tomlins AM, Ben-Rached F, Williams RA, Proto WR, Coppens I, Ruch U, Gilberger TW, Coombs GH, Mottram JC, Müller S, Langsley (2013) ''Plasmodium falciparum'' ATG8 implicated in both autophagy and apicoplast formation" ''Autophagy'' 9(10)</ref> The digestive vacuole is probably used for this purpose because the parasite lacks [[lysosome]]s. |
|||
===Crystaloids=== |
|||
These are transient structures whose presence is restricted to the mosquito specific ookinete and young oocyst stages of the parasite.<ref name=Dessens2011>Dessens JT, Saeed S, Tremp AZ, Carter V (2011) Malaria crystalloids: specialized structures for parasite transmission? Trends Parasitol 27(3) 106-110 {{DOI|10.1016/j.pt.2010.12.004}}</ref> They are cytoplasmic aggregations of closely packed spherical particles 25–35 nanometers in diameter and disappear after ookinete-to-oocyst transition. Although first described in the 1962 their function is unknown.<ref name=Garnham1962>Garnham PC, Bird RG, Baker JR (1962) Electron microscope studies of motile stages of malaria parasites. III. The ookinetes of ''Haemamoeba'' and ''Plasmodium''" ''Trans R Soc Trop Med Hyg'' 1962;56:116–120</ref> They contain a scavenger receptor like protein which is formed in the macrogametocytes.<ref name=Carter2008>Carter V, Shimizu S, Arai M, Dessens JT (2008) PbSR is synthesized in macrogametocytes and involved in formation of the malaria crystalloids" ''Mol Microbiol'' 68(6) 1560-1569 {{DOI|10.1111/j.1365-2958.2008.06254.x}}</ref> They also contain a number of LCCL proteins.<ref name=Saeed2010>Saeed S, Carter V, Tremp AZ, Dessens JT (2010) ''Plasmodium berghei'' crystalloids contain multiple LCCL proteins. Mol Biochem Parasitol 170(1) 49-53 {{DOI|10.1016/j.molbiopara.2009.11.008}}</ref> |
|||
===Effect of radiation=== |
|||
Gamma rays may be used to produce attenuated parasites. Despite this the effects on the parasite have rarely been studied.<ref name=Oakley2012>Oakley MS, Gerald N, Anantharaman V, Gao Y, Majam V, Mahajan B, Pham PT, Lotspeich-Cole L, Myers TG, McCutchan TF, Morris SL, Aravind L, Kumar S (2012) Radiation induced cellular and molecular alterations in asexual intraerythrocytic Plasmodium falciparum parasites. J Infect Dis</ref> Gamma irradation acts in a dose dependent fashion: morphologically it induces defective mitosis, sparse cytoplasm, fewer ribosomes, disorganized and clumped organelles and large vacuoles. The transcription of a number of genes is altered. |
|||
==Molecular biology and biochemistry== |
|||
===Redundancy of binding/invasion proteins=== |
|||
Proteins involved in this process are subject to three levels of host cell selection: |
|||
*selection between host species (species specific tropism) |
|||
*selection among individuals within a host species (erythrocyte receptor diversity) |
|||
*selection of subpopulations within an individual (age dependent invasion) |
|||
These proteins are necessarily surface exposed and are also subject to selection by the host's immune system |
|||
Because of the multiple levels of selection it is to be expected that a number of proteins are involved in this process. This also in part explains the apparent redundancy of these proteins. |
|||
===Surface exposed proteins=== |
|||
;Merozoite surface protein (MSP) family |
|||
The cleavage of MSP 1 appears to involve a purinergic signalling pathway.<ref name=daCruz2012>da Cruz LN, Juliano MA, Budu A, Juliano L, Holder AA, Blackman MJ, Garcia CR (2012) Extracellular ATP triggers proteolysis and cytosolic Ca2+ rise in Plasmodium berghei and Plasmodium yoelii malaria parasites. Malar J 11(1) 69.</ref> |
|||
The MSP1 protein binds the pro inflammatory protein [[S100P]].<ref name=Waisberg2012>Waisberg M, Cerqueira GC, Yager SB, Francischetti IM, Lu J, Gera N, Srinivasan P, Miura K, Rada B, Lukszo J, Barbian KD, Leto TL, Porcella SF, Narum DL, El-Sayed N, Miller LH, Pierce SK (2012) ''Plasmodium falciparum'' merozoite surface protein 1 blocks the proinflammatory protein S100P. Proc Natl Acad Sci USA</ref> This binding appears to prevent the usual NFκB activation in [[monocyte]]s and [[chemotaxis]] in [[neutrophil]]s. S100P appears to be able to bind to at least 2 alleles of MSP1 which are separated by at least 27 million years of evolution suggesting that this inhibition mechanism may also be of considerable age. |
|||
[[Merozoite surface protein]] 2 is one of the most abundant proteins on the surface of merozoites, is intrinsically unstructured and forms [[amyloid]]-like fibrils in solution. |
|||
Merozoite surface protein 7 appears to enhance the virulence of the parasite at least in the rodent.<ref name="Gómez2011">{{cite journal |last1=Gómez |first1=ND |last2=Safeukui |first2=I |last3=Adelani |first3=AA |last4=Tewari |first4=R |last5=Reddy |first5=JK |last6=Rao |first6=S |last7=Holder |first7=A |last8=Buffet |first8=P |last9=Mohandas |first9=N ''et al.'' |year=2011 |title=Deletion of a malaria invasion gene reduces death and anemia, in model hosts |journal=PLoS ONE |volume=6 |issue=9 |page=e25477 |doi=10.1371/journal.pone.0025477 |editor1-last=Spielmann |editor1-first=Tobias |last10=Haldar |first10=Kasturi}}</ref> |
|||
A protein PfMSPDBL1 (encoded by PF10_0348 gene) that is a member of the MSP3 family and has both Duffy binding-like (DBL) domain and secreted polymorphic antigen associated with merozoites (SPAM) domain appears to be critical for erythrocyte invasion.<ref name=Sakamoto2012>{{cite journal |author=Sakamoto H, Takeo S, Maier AG, Sattabongkot J, Cowman AF, Tsuboi T |year=2012 |title=Antibodies against a ''Plasmodium falciparum'' antigen PfMSPDBL1 inhibit merozoite invasion into human erythrocytes |journal=Vaccine |doi=10.1016/j.vaccine.2012.01.010 |volume=30 |issue=11 |pages=1972–80 |pmid=22248820}}</ref> The merozoite surface proteins DBL1 and -2 (PfMSPDBL1 and PfMSPDBL2) (PF10_0348 and PF10_0355) are extrinsically associated with the merozoite. MSPDBL2 appears to have a role in resistance to [[halofantrine]], [[mefloquine]] and [[lumefantrine]].<ref name=Van_Tyne2013>Van Tyne D, Uboldi AD, Healer J, Cowman AF, Wirth DF (2013) Modulation of PF10_0355 (MSPDBL2) alters ''Plasmodium falciparum'' response to antimalarial drugs. Antimicrob Agents Chemother</ref> |
|||
;Circumsporozoite protein (CSP) |
|||
The circumsporozoite protein (CSP) forms a dense coat on the sporozoite's surface.<ref>Huang YT, Lu XM, Jin XB, Zhu JY (2012) Research advances on circumsporzoite protein of ''Plasmodium''. Zhongguo Ji Sheng Chong Xue Yu Ji Sheng Chong Bing Za Zhi 30(3) 238-242</ref> It consists of approximately 400 amino acids organized into three domains: an N-terminal domain containing a conserved pentapeptide (region I), a highly repetitive species specific central domain and a C-terminal domain containing a second conserved sequence (region II). It is involved in invasion of the mosquito's salivary glands and the binding sporozoites to liver cells. |
|||
The circumsporozoite protein has been shown to be an inhibitor of the nuclear factor kappa-light-chain-enhancer of activated B cells ([[NF-κB]]).<ref name=Ding2012>Ding Y, Huang X, Liu T, Fu Y, Tan Z, Zheng H, Zhou T, Dai J, Xu W (2012) The ''Plasmodium'' circumsporozoite protein, a novel NF-κB inhibitor, suppresses the growth of SW480. Pathol Oncol Res</ref> Its nuclear localization signal alone is sufficient to block NF-κB activation. |
|||
CSP binds to salivary glands and is involved in its invasionIn by the sporozoites. An Anopheles salivary gland protein - CSP-binding protein - to which CSP binds during this process has been identified.<ref name=Wang2013>Wang J, Zhang Y, Zhao YO, Li MW, Zhang L, Dragovic S, Abraham NM, Fikrig E (2013) ''Anopheles gambiae'' circumsporozoite-protein binding-protein facilitates ''Plasmodium'' infection of mosquito salivary glands. J Infect Dis</ref> |
|||
;Early transcribed membrane protein (ETRAMP) family |
|||
The ETRAMP family is characterized by a predicted [[signal peptide]], a short [[lysine]] rich stretch, an internal transmembrane domain and a highly charged C-terminal region of variable length. The highly charged terminal region appears to be involved in protein-protein interactions.<ref name="Currà2011">Currà C, Pace T, Franke-Fayard BM, Picci L, Bertuccini L, Ponzi M (2011) Erythrocyte remodeling in ''Plasmodium berghei'' infection: the contribution of SEP family members. Traffic {{doi|10.1111/j.1600-0854.2011.01313.x}}</ref> They are usually expressed in a stage-specific manner. In the blood stages they localize to the parasitophorous vacuole membrane and to vesicle like structures exported to the host erythrocyte cytosol. The gene ETRAMP 10.3 has been shown to be expressed in the liver, sporozoites and blood stages.<ref name=Mackellar2010>Mackellar DC, O'Neill MT, Aly AS, Sacci JB Jr, Cowman AF, Kappe SH (2010) ''Plasmodium falciparum'' PF10_0164 (ETRAMP10.3) is an essential parasitophorous vacuole and exported protein of blood stages. Eukaryotic Cell</ref> Within the liver and blood stages it is localized to the parasitophorous vacuole membrane. It is also exported to the erythrocyte during the blood stages. It appears to be an essential gene in the blood stages.<ref name=Mackellar2011>Mackellar DC, Vaughan AM, Aly AS, Deleon S, Kappe SH (2011) A systematic analysis of the early transcribed membrane protein family throughout the life cycle of ''Plasmodium yoelii''. Cell Microbiol. {{doi|10.1111/j.1462-5822.2011.01656.x}}</ref> In ''[[Plasmodium berghei]]'' two members of the ETRAMP family (uis3 and uis4) localize to secretory organelles of sporozoites and to the parasitophorous membrane vacuole of the liver stages.<ref name="Currà2013">Currà C, Di Luca M, Picci L, de Sousa Silva Gomes Dos Santos C, Siden-Kiamos I, Pace T, Ponzi M (2013) The ETRAMP family member SEP2 is expressed throughout ''Plasmodium berghei'' life cycle and is released during sporozoite gliding motility" ''PLoS One'' 8(6) e67238. {{DOI|10.1371/journal.pone.0067238}}</ref> Another member of this family - SEP2 - is expressed in the gametocytes, in the mosquito and in the liver stages.<ref name="Currà2013"/> In the liver stage SEP2 is routed to the parasitophorous vacuole membrane. In the ookinete and sporozoite stages, it instead localizes to the parasite surface. It is also released during gliding motility of salivary gland sporozoites. |
|||
;6-cys domain proteins |
|||
A group of proteins known as the 6-cys domain proteins - so called because they contain modules with six characteristic cysteines forming three intra-molecular disulphide bonds between C1 and C2, C3 and C6, and C4 and C5 - are surface exposed proteins.<ref name=Taechalertpaisarn2012>Taechalertpaisarn T, Crosnier C, Bartholdson SJ, Hodder AN, Thompson J, Bustamante LY, Wilson DW, Sanders PR, Wright GJ, Rayner JC, Cowman AF, Gilson PR, Crabb BS (2012) Biochemical and functional analysis of two ''Plasmodium falciparum'' blood-stage 6-cys proteins: P12 and P41" ''PLoS One'' 7(7) e41937.</ref> The first P12 - named after the clone it was isolated from - was described in 1990.<ref name=Elliott1990>Elliott JF, Albrecht GR, Gilladoga A, Handunnetti SM, Neequaye J, Lallinger G, Minjas JN, Howard RJ (1990) Genes for ''Plasmodium falciparum'' surface antigens cloned by expression in COS cells" ''Proc Natl Acad Sci USA'' 87(16) 6363-6367</ref> There are at least nine members of the 6-cys family. Most family members contain two 6-cys modules, but up to seven modules can be found in a single protein, in addition to incomplete modules containing fewer cysteine residues. About half of the 6-cys family members characterised to date possess glycosylphosphatidylinositol (GPI) moieties that anchor them to the outer leaflet of the plasma membrane, while those that lack GPI-anchors presumably remain associated with the parasite surface via interactions with other membrane proteins. Of this family P12, P38 and P41 are blood stage antigens. P230 and P48/45 - another two members of this family - are expressed on the surface of gametes. |
|||
P12 has only two s48/45 domains while other members have up to fourteen.<ref name=Tonkin2013>Tonkin ML, Arredondo SA, Loveless BC, Serpa JJ, Makepeace KA, Sundar N, Petrotchenko EV, Miller LH, Grigg ME, Boulanger MJ (2013) Structural and biochemical characterization of ''Plasmodium falciparum'' 12 (Pf12) reveals a unique inter-domain organization and the potential for an antiparallel arrangement with Pf41. J Biol Chem</ref> Pf12 is highly conserved and under purifying selection. It forms a heterodimeric complex with Pf41. |
|||
;LCCL/lectin adhesive-like protein family |
|||
There are a family of LCCL/lectin adhesive-like protein (LAP) proteins encoded in the genome.<ref name=Saeed2012>Saeed S, Tremp AZ, Dessens JT (2012) Conformational co-dependence between ''Plasmodium berghei'' LCCL proteins promotes complex formation and stability. Mol Biochem Parasitol</ref> The six members are expressed in gametocytes and form a multi-protein complex. There are normally six of these proteins in the genome. They are essential for parasite transmission to the mosquito.<ref name=Saeed2013>Saeed S, Carter V, Tremp AZ, Dessens JT (2013) Translational repression controls temporal expression of the Plasmodium berghei LCCL protein complex. Mol Biochem Parasitol pii: S0166-6851(13)00053-4. {{DOI|10.1016/j.molbiopara.2013.04.006}}</ref> |
|||
;Pf332 protein |
|||
The 700 kiloDalton protein Pf332 is the largest known exported asexual malaria protein. The protein has three parts: an N-terminal Duffy binding like domain followed by a putative transmembrane region and a large number of negatively charged repeats that are not identical but have the consensus (X)<sub>3</sub>-EE-(X)<sub>2</sub>-EE-(X)<sub>2–3</sub> where E is [[glutamic acid]] and X is a hydrophobic amino acid. The repeat portion of the protein consititue more than 90% of the protein. The protein has a predicted isoelectric point (pI) of 3.8. It is known to associate with the erythrocyte plasma membrane.<ref name=Mattei1992>Mattei D, Scherf A (1992) The Pf332 gene codes for a megadalton protein of ''Plasmodium falciparum'' asexual blood stages. Mem Inst Oswaldo Cruz 87: 163–168</ref><ref name=Moll2997>Moll K, Chene A, Ribacke U, Kaneko O, Nilsson S ''et al'' (2007) A novel DBL-domain of the ''P. falciparum'' 332 molecule possibly involved in erythrocyte adhesion" ''PLoS ONE'' 2: e477</ref> |
|||
The Pf332 protein can first be detected within the parasite at 20–24 hours post invasion, after which it translocates across the parasitotopherous vacuole membrane into the host cell cytosol.<ref name=Hinterberg1994>Hinterberg K, Scherf A, Gysin J, Toyoshima T, Aikawa M ''et al'' (1994) ''Plasmodium falciparum'': the Pf332 antigen is secreted from the parasite by a brefeldin A dependent pathway and is translocated to the erythrocyte membrane via the Maurer's clefts. Exp Parasitol 79: 279–291</ref> It is initially synthesised in the endoplasmic reticulum and eported to the host cytosol. From there it is trafficked as part of a multimeric protein complex to Maurer's clefts. It may interact with two chaperone proteins - PF14_0700 (a hypothetical protein with a J domain) and PFB0595w (a [[heat shock protein]] 40).<ref name=Pavithra2007>Pavithra SR, Kumar R, Tatu U (2007) Systems analysis of chaperone networks in the malarial parasite ''Plasmodium falciparum''" ''PLoS Comput Biol'' 3: 1701–1715</ref> It is associated with the cytoplasmic side of Maurer's clefts in a peripheral manner throughout trophozoite maturation and schizogony.<ref name=Nilsson2012>Nilsson S, Angeletti D, Wahlgren M, Chen Q, Moll K (2012) ''Plasmodium falciparum'' antigen 332 is a resident peripheral membrane protein of Maurer's clefts" ''PLoS One'' 7(11) e46980. {{DOI|10.1371/journal.pone.0046980}}</ref> In the clefts both the N and C-termini are localised to the erythocyte cytosol.<ref name="Nilsson2012"/> Export of Pf332 is sensitive to treatment with [[Brefeldin A]]<ref name="Hinterberg1994"/> The export signal appears to be encoded in the N terminal domain<ref name=Hodder2009>Hodder AN, Maier AG, Rug M, Brown M, Hommel M ''et al'' (2009) Analysis of structure and function of the giant protein Pf332 in ''Plasmodium falciparum''" ''Mol Microbiol'' 71: 48–65</ref> |
|||
It interacts with the erythrocyte cytoskeleton and binds actin.<ref name="Glenister2009"/><ref name=Waller2010>Waller KL, Stubberfield LM, Dubljevic V, Buckingham DW, Mohandas N ''et al'' (2010) Interaction of the exported malaria protein Pf332 with the red blood cell membrane skeleton" ''Biochim Biophys Acta'' 1798: 861–871 {{DOI|10.1016/j.bbamem.2010.01.018}}</ref> |
|||
;PHIST family |
|||
The ''Plasmodium'' helical intersperse sub-telomeric family is a collection of 72 small exported proteins.<ref name=Prajapati2013>Prajapati SK, Singh OP (2013) Remodeling of human red cells infected with Plasmodium falciparum and the impact of PHIST proteins. Blood Cells Mol Dis pii: S1079-9796(13)00149-6. {{DOI|10.1016/j.bcmd.2013.06.003}}</ref> |
|||
;Other proteins |
|||
The merozoite specific thrombospondin related anonymous protein (MTRAP) is thought to be released from the micronemes during merozoite invasion and mediates motility and host cell invasion through an interaction with aldolase.<ref name="Uchime2012"/> MTRAP is a highly extended bifunctional protein that binds to an erythrocyte receptor and the merozoite motor. MTRAP specific antibodies fail to inhibit parasite development ''in vitro''. |
|||
Thrombospondin related apical membrane protein (PTRAMP) is a surface exposed protein whose function is currently unknown.<ref name="Uchime2012"/> |
|||
The phosphoprotein [[RPLP0|P0]] is surface exposed during the asexual erythrocytic stages and [[antibody|antibodies]] to this protein appear to be protective.<ref name=Das2012/> It is also present on the surface of the merozoites. |
|||
The [[RPLP2|60S stalk ribosomal acidic protein P2]] (gene PFC0400w) as well as forming part of the ribosome complex is surface exposed where it forms homo-tetramers.<ref name=Das2012/> This protein is exported to the erythrocyte surface 26-28 post invasion and persists there for 6–8 hours. Treatment with antiP2 antibodies causes mitotic arrest at the first nuclear division and disruption of the tubovesicular network which is set up during the trophozoite stages. Removal of the antibodies al lows the reformation of the tubovesicular network and mitotic division to continue. |
|||
The receptor for the attachment protein PfRh4 has been identified as [[complement receptor 1]].<ref name="Tham2010">{{cite journal |last1=Tham |first1=WH |last2=Wilson |first2=DW |last3=Lopaticki |first3=S |last4=Schmidt |first4=CQ |last5=Tetteh-Quarcoo |first5=PB |last6=Barlow |first6=PN |last7=Richard |first7=D |last8=Corbin |first8=JE |last9=Beeson |first9=JG ''et al.'' |year=2010 |title=Complement receptor 1 is the host erythrocyte receptor for Plasmodium falciparum PfRh4 invasion ligand |journal=Proc Natl Acad Sci U S A |volume=107 |issue=40 |pages=17327–17332 |display-authors=9 |doi=10.1073/pnas.1008151107 |pmid=20855594 |pmc=2951459 |last10=Cowman |first10=A. F.}}</ref> |
|||
The [[mature parasite-infected erythrocyte surface antigen]] (MESA) is exported to the erythrocyte cytoplasm where it binds to the N-terminal 30 kilo[[Dalton (unit)|Dalton]] domain of the erythrocyte protein 4.1R via a 19-residue sequence.<ref name=Kilili2011>{{cite journal |author=Kilili GK, Lacount DJ |year=2011 |title=An erythrocyte cytoskeleton-binding motif in exported ''Plasmodium falciparum'' proteins |journal=[[Eukaryotic Cell]] |doi=10.1128/EC.05180-11 |volume=10 |issue=11 |pages=1439}}</ref> This sequence is also found in a number of other proteins in the parasite. Their role in remodeling of the erythrocyte are still under investigation. |
|||
The Ring-Infected Erythrocyte Surface Antigen (RESA/Pf155) protein appears to affect the mobility of the erythrocyte membrane.<ref name=Diez-Silva2012>Diez-Silva M, Park Y, Huang S, Bow H, Mercereau-Puijalon O, Deplaine G, Lavazec C, Perrot S, Bonnefoy S, Feld MS, Han J, Dao M, Suresh S (2012) Pf155/RESA protein influences the dynamic microcirculatory behavior of ring-stage Plasmodium falciparum infected red blood cells. Sci Rep 2:614</ref> |
|||
The proteins Pf12, Pf34, Pf92 and Pf38 are associated with detergent resistant membrane microdomains through glycosylphosphatidylinositol anchor sequences.<ref name=Gilson2006>Gilson PR, Nebl T, Vukcevic D, Moritz RL, Sargeant T, Speed TP, Schofield L, Crabb BS (2006) Identification and stoichiometry of glycosylphosphatidylinositolanchored membrane proteins of the human malaria parasite ''Plasmodium falciparum''. Mol Cell Proteomics 5:1286-1299</ref> These microdomains are considered organizing centers for the assembly of molecules implicated in cell signaling. |
|||
The erythrocyte binding like 175 protein (EBL) is normally found in the micronemes. It has a signal sequence and a [[cysteine]] rich domain both of which are required for localisation to the micronene. In addition to these two ssequences for correct trafficking it also requires a sequence in its region 5.<ref name=Sakura2012>Sakura T, Yahata K, Kaneko O (2012) The upstream sequence segment of the C-terminal cysteine-rich domain is required for microneme trafficking of ''Plasmodium falciparum'' erythrocyte binding antigen 175. Parasitol Int. 2012 Dec 22. pii: S1383-5769(12)00162-6. {{DOI|10.1016/j.parint.2012.12.002}}</ref> |
|||
[[Enolase]] is bound to the surface of ''P. falciparum'' and several other pathogens.<ref name=Ghosh2011>Ghosh AK, Jacobs-Lorena M (2011) Surface-expressed enolases of ''Plasmodium'' and other pathogens. Mem Inst Oswaldo Cruz 106(Suppl 1) 85-90</ref> In this location it binds [[plasminogen]] which is thought to function in the degradation of the extracellular matrix surrounding the targeted host cell, thereby facilitating pathogen invasion. |
|||
The gene PFE0565w is transcribed in both the erythrocytic and sporozoite stages.<ref name=Schlarman2012>{{cite journal |last1=Schlarman |first1=MS |last2=Roberts |first2=RN |last3=Kariuki |first3=MM |last4=Lacrue |first4=AN |last5=Ou |first5=R |last6=Beerntsen |first6=BT |year=2012 |title=PFE0565w, a ''Plasmodium falciparum'' protein Expressed in salivary gland sporozoites |journal=Am J Trop Med Hyg |volume=86 |issue=6 |pages=943–954 |doi=10.4269/ajtmh.2012.11-0797 |pmid=22665598 |pmc=3366537}}</ref> The protein is only expressed in the salivary gland sporozoite stage. |
|||
Surfin 4.1 - a type I transmembrane protein located on the merozoite surface - is responsible for reversible adherence to the erythrocyte before invasion.<ref name=Xangsayarath2012>Xangsayarath P, Kaewthamasorn M, Yahata K, Nakazawa S, Sattabongkot J, Udomsangpetch R, Kaneko O (2012) Positive diversifying selection on the ''Plasmodium falciparum'' surf gene in Thailand. Trop Med Health 40(3) 79-89 {{DOI|10.2149/tmh.2012-12}}</ref> The gene is highly polymorphic, particularly at the C-terminal side of the variable region located just before a predicted transmembrane region. Positive diversifying selection is detectable in this region and in the conserved N-terminally located cysteine-rich domain. |
|||
[[Heparin]] has been shown to bind to infected erythrocytes.<ref name=Valle-Delgado2012>Valle-Delgado JJ, Urbán P, Fernàndez-Busquets X (2013) Demonstration of specific binding of heparin to Plasmodium falciparum-infected vs. non-infected red blood cells by single-molecule force spectroscopy. Nanoscale</ref> What role this binding has in the pathology of this infection - if any - is not yet clear. |
|||
===Erythrocyte proteins taken up=== |
|||
A small number of erythrocytic proteins are taken up by the parasite during the course of its life cycle. The role these play is not clear. Among these proteins is [[dematin]] which interacts with the parasite's [[14-3-3]] protein.<ref name="Lalle2010">{{cite journal |author=Lalle M, Curra C, Ciccarone F, Pace T, Cecchetti S, Fantozzi L, Ay B, Braun Breton C, Ponzi M. |year=2010 |title=Dematin, a component of the erythrocyte membrane-skeleton, is internalized by the malaria parasite and associates with ''Plasmodium'' 14-3-3 |journal=J. Biol. Chem.}}</ref> |
|||
The parasite is capable of making use of the erythrocyte's own enzymes. The enzymes [[PAK1]] and [[MEK1]] neither of which are encoded in the ''Plasmodium'' genome have been shown to be phosphorylated and activated during the course of infection<ref name="Sicard2011">{{cite journal |author=Sicard A, Semblat JP, Doerig C, Hamelin R, Moniatte M, Dorin-Semblat D, Spicer JA, Srivastava A, Retzlaff S, Heussler V, Waters AP, Doerig C |year=2011 |title=Activation of a PAK-MEK signalling pathway in malaria parasite-infected erythrocytes |journal=Cell Microbiol. |doi=10.1111/j.1462-5822.2011.01582.x |volume=13 |issue=6 |pages=836–45 |pmid=21371233 |pmc=3123749}}</ref>' ''In vitro'' work has shown that inhibition of these enzymes is fatal to the parasite. |
|||
''Plasmodium'' ingests [[kininogen]] from which its proteases generate vasoactive peptides.<ref name=Bagnaresi2012>Bagnaresi P, de Barros NM, Assis DM, Melo PM, Fonseca RG, Juliano MA, Pesquero JB, Juliano L, Rosenthal PJ, Carmona AK, Gazarini ML (2012) Intracellular proteolysis of kininogen by malaria parasites promotes release of active kinins. Malar J 11(1) 156</ref> The role this may play in the pathophysiology of malaria is not yet understood. |
|||
Another enzyme that is imported by the parasite is the human redox-active protein peroxiredoxin 2 (hPrx-2, hTPx1).<ref name=Koncarevic2009>Koncarevic S, Rohrbach P, Deponte M, Krohne G, Prieto JH, Yates J 3rd, Rahlfs S, Becker K (2009) The malarial parasite ''Plasmodium falciparum'' imports the human protein peroxiredoxin 2 for peroxide detoxification" ''Proc Natl Acad Sci USA'' 106(32) 13323-8. {{DOI|10.1073/pnas.0905387106}}</ref> This imported protein accounts for ~50% of the total [[thioredoxin peroxidase]] activity in parasite extracts. In the presence of chloroquine the parasite increases its imports of this protein. This protein is found both in the cytosol and in Maurer's clefts. |
|||
Another enzyme that is taken up is [[delta-aminolevulinate dehydrase]] - an enzyme in the haem biosynthesis pathway.<ref name=Bonday1997>Bonday ZQ, Taketani S, Gupta PD, Padmanaban G (1997) Heme biosynthesis by the malarial parasite. Import of delta-aminolevulinate dehydrase from the host red cell" ''J Biol Chem'' 272(35) 21839-21846</ref> |
|||
===Transport/secretion=== |
|||
The uninfected erythrocyte lacks a regulated transport system. Vesicular transport within both the parasite and the infected erythrocyte cytoplasm must be provided by the parasite itself. |
|||
Both the cytoplasmic pH (7.3) and the inside negative plasma membrane potential (-95 mV) are kept fairly constant during the intra erythrocytic cycle. This is due to the action of a V-type H(+)-ATPase which is also responsible for the pH of the digestive vacuole. There is also a Na<sup>+</sup> ATPase in the plasma membrane.<ref name=Spillman2013>Spillman NJ, Allen RJ, Kirk K (2013) Na+ extrusion imposes an 'acid load' on the intraerythrocytic malaria parasite. Mol Biochem Parasitol. 2013 Apr 23. pii: S0166-6851(13)00051-0. {{DOI|10.1016/j.molbiopara.2013.04.004}}</ref> |
|||
;Transport |
|||
The intracellular concentration of [[chloride]] ions has been estimated to be 48 milliMolar.<ref name="Henry2010">{{cite journal |author=Henry RI, Cobbold SA, Allen RJ, Khan A, Hayward R, Lehane AM, Bray PG, Howitt SM, Biagini GA, Saliba KJ, Kirk K |year=2010 |title=An acid-loading chloride transport pathway in the intraerythrocytic malaria parasite, ''Plasmodium falciparum'' |journal=J. Biol. Chem. |doi=10.1074/jbc.M110.120980 |volume=285 |issue=24 |pages=18615–26 |pmid=20332090 |pmc=2881787}}</ref> It appears to actively import using [[Adenosine triphosphate|ATP]] both [[hydrogen]] [[ion]]s and [[chloride]] ions in a linked fashion via a DIDS sensitive transporter in the cytoplasmic membrane. |
|||
One difficulty the parasite has in acquiring nutrients from the cytoplasm is the presence of phosphate groups on these molecules. It appears to have overcome this by secreting an acid phosphatase ([[glideosome-associated protein 50]] - GAP50 ) into the cytoplasm that is then taken up into the digestive vacuole.<ref name="Müller2010">Müller IB, Knöckel J, Eschbach ML, Bergmann B, Walter RD, Wrenger C (2010) Secretion of an acid phosphatase provides a possible mechanism to acquire) host nutrients by ''Plasmodium falciparum''. Cell Microbiol.</ref> |
|||
The parasite has an absolute requirement for [[isoleucine]] - an [[amino acid]] absent from human [[haemoglobin]]. A saturable neutral amino acid ([[methionine]], [[leucine]], isoleucine) transporter appears to be encoded by the parasite and this protein functions in the infected erythrocyte membrane.<ref name="Cobbold2010">{{cite journal |author=Cobbold SA, Martin RE, Kirk K |year=2010 |title=Methionine transport in the malaria parasite ''Plasmodium falciparum'' |journal=Parasitol.}}</ref> |
|||
The ''P. falciparum'' Na+/H+ exchanger (PfNHE1) is located on chromosome 13 (gene PF13_0019).<ref name=Sinou2011>Sinou V, Quang LH, Pelleau S, Huong VN, Huong NT, Tai LM, Bertaux L, Desbordes M, Latour C, Long LQ, Thanh NX, Parzy D (2011) Polymorphism of ''Plasmodium falciparum'' Na+/H+ exchanger is indicative of a low in vitro quinine susceptibility in isolates from Viet Nam. Malar J. 2011 Jun 14;10(1) 164</ref> This gene may be involved in resistance to quinine.<ref name=Kone2012>Kone A, Mu J, Maiga H, Beavogui AA, Yattara O, Sagara I, Tekete MM, Traore OB, Dara A, Dama S, Diallo N, Kodio A, Traoré A, Björkman A, Gil JP, Doumbo OK, Wellems TE, Djimde AA (2012) Quinine treatment selects Pfnhe1 ms47601 polymorphism in Malian patients with falciparum malaria. J Infect Dis</ref> |
|||
Two folate transporters (PfFT1 and PfTF2) have been cloned.<ref name=Salcedo-Sora2011>{{cite journal |author=Salcedo-Sora JE, Ochong E, Beveridge S, Johnson D, Nzila A, Biagini GA, Stocks PA, O'Neill PM, Krishna S, Bray PG, Ward SA |year=2011 |title=The molecular basis of folate salvage in ''Plasmodium falciparum'': Characterization of two folate transporters |journal=J Biol Chem |doi=10.1074/jbc.M111.286054 |volume=286 |issue=52 |pages=44659–68 |pmid=21998306 |pmc=3247980}}</ref> Substrates include [[folic acid]], [[folinic acid]], the folate precursor pABA and the human folate catabolite pABAG(n). 5-methyl tetrahydofolate is not transported by PfFT1 and only poorly by PfFT2. The activity of both transporters may be inhibited by [[probenecid]] or [[methotrexate]]. Folate transport appears to be an ATP requiring activity and dependent on a proton gradient.<ref name=Wang2009>Wang P, Wang Q, Sims PF, Hyde JE (2009) Characterisation of exogenous folate transport in ''Plasmodium falciparum''. Mol Biochem Parasitol 154(1) 40-51</ref> |
|||
The parasite possesses its own [[equilibrative nucleoside transporter]] 1. All members of this protein family have 11 transmembrane segments. The gene product is located in the parasite's plasma membrane and knock out mutants have shown that this is an essential gene at least at physiological concentrations. In the 11th transmembrane segment two mutations have been shown to affect its activity: a [[phenylalanine]] (Phe) to [[leucine]] (Leu) at residue 394 (F394L) via [[cytosine]] (C) or [[uracil]] (U) to [[adenosine]] (A) or [[guanine]] (G) at the third [[codon]] position and a [[cysteine]] (Cys) to [[glycine]] (Gly) mutation at either glycine in a conserved glycine-X-X-glycine motif (where X is any amino acid) via a cytosine to uracil at the second codon position.<ref name="Riegelhaupt2010">Riegelhaupt PM, Frame IJ, Akabas MH (2010) Transmembrane segment 11 appears to line the purine permeation pathway of the Plasmodium falciparum equilibrative nucleoside transporter 1 (PfENT1). J Biol Chem.</ref> Additional work suggests that the 11th transmembrane segment is largely alpha helical. It has been suggested that this transmembrane segment may be the actual purine transport channel. |
|||
The parasite is unable to synthesize purines (including [[adenosine]], [[hypoxanthine]] and [[adenine]]) and must take these up from the host. Purines are transported across the parasite plasma membrane entry into the infected erythrocyte ''P. falciparum'' nucleoside transporter 1 (PfNT1).<ref name=ElBissati2006>El Bissati K, Zufferey R, Witola WH, Carter NS, Ullman B, Ben Mamoun C (2006) The plasma membrane permease PfNT1 is essential for purine salvage in the human malaria parasite ''Plasmodium falciparum''" ''Proc Natl Acad Sci USA'' 103(24) 9286-9291</ref> This transport system carries [[hypoxanthine]], [[inosine]] and [[adenosine]] into the parasite. At least some of the hypoxanthine is converted into uric acid by the parasite. |
|||
Three purine transporters have been studied: the human [[equilibrative nucleoside transporter]] (hENT1), the human [[facilitative nucleobase transporter]] (hFNT1) and the parasite-induced new permeation pathway (NPP). The bulk of transport is facilitated by host's own transporters rather than through the NPP.<ref name=Quashie2010>Quashie NB, Ranford-Cartwright LC, De Koning HP (2010) Uptake of purines in ''Plasmodium falciparum''-infected human erythrocytes is mostly mediated by the human equilibrative nucleoside transporter and the human Facilitative Nucleobase Transporter. Malar J 9(1) 36</ref> Hypoxanthine and adenine were transported mainly through the hFNT1 pathway whereas adenosine entered predominantly through the hENT1 system. The rate of purine uptake in infected cells was approximately twice that of uninfected erythrocytes. The rate of adenosine uptake was greater than the rate of hypoxanthine uptake in infected human red blood cells. [[Furosemide]] inhibits the transport of purine bases through the hFNT1. |
|||
An intracellular [[purine]] [[permease]] (PfNT2) has been shown to be localised to the [[endoplasmic reticulum]].<ref name="Downie2010">{{cite journal |author=Downie MJ, El Bissati K, Bobenchik AM, Nic Lochlainn L, Amerik A, Zufferey R, Kirk K, Ben Mamoun C |year=2010 |title=PfNT2: a permease of the equilibrative nucleoside transporter family in the endoplasmic reticulum of ''Plasmodium falciparum'' |journal=J Biol Chem. |doi=10.1074/jbc.M110.118489 |volume=285 |issue=27 |pages=20827–33 |pmid=20439460 |pmc=2898299}}</ref> This protein is a member of the equilibrative nucleoside transporter family. |
|||
Within the genome there are encoded four equilibrative nucleoside transporters (ENTs). ENT 1 is the major route of purine nucleoside/nucleobase transport in the erythrocytic stages. Knock out mutants have been generated that can survive. ENT4 has been cloned and expressed.<ref name=Frame2012>Frame IJ, Merino EF, Schramm VL, Cassera MB, Akabas MH (2012) Malaria parasite type 4 equilibrative nucleoside transporters (ENT4) are purine transporters with distinct substrate specificity. Biochem J</ref> It does not appear to transport either [[hypoxanthine]] or [[adenine monophosphate]] but does transport [[adenine]] and 2'-deoxyadenosine. It is inhibited by [[dipyridamole]]. |
|||
The parasite can uptake polyamines from the host. Two of these - [[putrescine]] and [[spermidine]] - are taken up in a temperature, pH and membrane potential dependent mechanism.<ref name=Niemand2012>Niemand J, Louw AI, Birkholtz L, Kirk K (2012) Polyamine uptake by the intraerythrocytic malaria parasite, ''Plasmodium falciparum''. Int J Parasitol</ref> |
|||
The clag3 genes on chromosome 3 appear to be involved in anion transport rather than in cell adherence as originally thought.<ref name=Nguitragool2011>{{cite journal |author=Nguitragool W, Bokhari AA, Pillai AD, Rayavara K, Sharma P, Turpin B, Aravind L, Desai S |year=2011 |title=Malaria parasite clag genes determine nutrient uptake channel activity on infected red blood cells |journal=Cell |volume=145 |issue=5 |pages=665–677 |doi=10.1016/j.cell.2011.05.002 |pmid=21620134 |pmc=3105333}}</ref> |
|||
At least two of the ''clag3'' genes appear to be involved in the surface anion channel which functions in nutrient uptake.<ref name=Pillai2012>Pillai AD, Nguitragool W, Lyko B, Dolinta K, Butler MM, Nguyen ST, Peet NP, Bowlin TL, Desai SA (2012) Solute restriction reveals an essential role for clag3-associated channels in malaria parasite nutrient acquisition. Mol Pharmacol</ref> |
|||
The clag3 gene family encode a parasite ion channel known as the plasmodial surface anion channel. Its activation appears to involve an intracellular domain.<ref name=Alkhalil2011>{{cite journal |author=Alkhalil A, Hong L, Nguitragool W, Desai SA |year=2011 |title=Voltage-dependent inactivation of the plasmodial surface anion channel via a cleavable cytoplasmic component |journal=Biochim Biophys Acta}}</ref> The anion channel is formed by 5 clag genes and forms a complex with RhopH and other proteins.<ref name=Alexandre2012>Alexandre JS, Xangsayarath P, Kaewthamasorn M, Yahata K, Sattabongkot J, Udomsangpetch R, Kaneko O (2012) Stable allele frequency distribution of the ''Plasmodium falciparum'' ''clag'' genes encoding components of the high molecular weight rhoptry protein complex. Trop Med Health 40(3) 71-77 {{DOI|10.2149/tmh.2012-13}}</ref> The expression of the anion channel can be suppressed by reversible histone modification.<ref name=Sharma2013>Sharma P, Wollenberg K, Sellers M, Zainabadi K, Galinsky K, Moss E, Nguitragool W, Neafsey D, Desai SA (2013) An epigenetic antimalarial resistance mechanism involving parasite genes linked to nutrient uptake. J Biol Chem</ref> |
|||
Two paralogous clag3 genes - clag3.1 and clag3.2 - show mutually exclusive expression and can be silenced by epigenetic mechanisms.<ref name="Mira-Martínez2013">Mira-Martínez S, Rovira-Graells N, Crowley VM, Altenhofen LM, Llinás M, Cortés A (2013) Epigenetic switches in clag3 genes mediate blasticidin S resistance in malaria parasites. Cell Microbiol {{DOI|10.1111/cmi.12162}}</ref> They appear to offer different transport efficiency for some solutes. The expression of at least one of these genes is essential for the function of the anion channel. |
|||
Positive diversifying selection has acted upon clag2, clag8 and clag9 but not in clag3.1 and clag3.2.<ref name=Alexandre2011>Alexandre JS, Kaewthamasorn M, Yahata K, Nakazawa S, Kaneko O (2011) Positive selection on the ''Plasmodium falciparum'' clag2 gene encoding a component of the erythrocyte-binding rhoptry protein complex. Trop Med Health 39(3) 77-82</ref> These proteins appear to be involved in anion transport. |
|||
The plasma membrane protein [[aquaglyceroporin]] mediates the transport of both [[glycerol]] and water.<ref name=Chen2012>Chen LY (2012) Glycerol inhibits water permeation through ''Plasmodium falciparum'' aquaglyceroporin. J Struct Biol pii: S1047-8477(12)00289-4. {{DOI|10.1016/j.jsb.2012.10.007}}</ref> |
|||
A [[copper]] transport protein (PF14_0369) has been identified<ref name=Choveaux2012>Choveaux DL, Przyborski JM, Goldring JD (2012) A ''Plasmodium falciparum'' copper-binding membrane protein with copper transport motifs. Malar J 11(1) 397</ref> This protein is expressed in early ring stage and translocating from the erythrocyte plasma membrane to a parasite membrane as the parasites developed to schizonts. Inhibition of copper uptake with [[neocuproine]] inhibits the ring to trophozoite transition. |
|||
The parasite is dependent on the acquisition of [[pantothenate]] from the host. The transporter - PfPAT - has been cloned.<ref>Augagneur Y, Jaubert L, Schiovani M, Pachikara N, Garg A, Usmani-Brown S, Wesolowski D, Zeller S, Ghosal A, Cornillot E, Said HM, Kumar P, Altman S, Ben Mamoun C (2013) Identification and functional analysis of the primary pantothenate transporter, PfPAT, of the human malaria parasite ''Plasmodium falciparum''. J Biol Chem</ref> The transporter is located in the parasite's plasma membrane and plays an essential role in its intraerythrocytic development. It can be inhibited by the drug [[fenpropimorph]]. |
|||
;Secretion |
|||
Protein export into the infected erythrocyte is critical for malaria parasite survival and the majority of effector proteins are thought to export via a proteinaceous translocon.<ref name=Riglar2013>Riglar DT, Rogers KL, Hanssen E, Turnbull L, Bullen HE, Charnaud SC, Przyborski J, Gilson PR, Whitchurch CB, Crabb BS, Baum J, Cowman AF (2013) Spatial association with PTEX complexes defines regions for effector export into ''Plasmodium falciparum''-infected erythrocytes. Nat Commun 4:1415. {{DOI|10.1038/ncomms2449}}</ref> This is found in the parasitophorous vacuole membrane surrounding the parasite. Exported protein 2 is a critical component of this system. |
|||
A translocon (PTEX) of several proteins is located in the vacuole membrane.<ref name=deKoning-Ward2009>de Koning-Ward TF, Gilson PR, Boddey JA, Rug M, Smith BJ, Papenfuss AT, Sanders PR, Lundie RJ, Maier AG, Cowman AF, Crabb BS (2009) A newly discovered protein export machine in malaria parasites" ''Nature'' 459(7249) 945-9. {{DOI|10.1038/nature08104}}</ref> These include heat shock protein 101 (HSP101) a ClpA/B-like ATPase from the AAA+ superfamily, of a type commonly associated with protein translocons, a novel protein termed PTEX150 and a known parasite protein - exported protein 2 (Exp2). Exp2 is the potential channel as it is the membrane-associated component of the core PTEX complex. Two other proteins - a new protein PTEX88 and thioredoxin 2 (Trx2) - are also PTEX components. These latter two proteins do not appear to be essential.<ref name=Matz2013>Matz JM, Matuschewski K, Kooij TW (2013) Two putative protein export regulators promote malaria blood stage development ''in vivo''. Mol Biochem Parasitol pii: S0166-6851(13)00134-5. {{DOI|10.1016/j.molbiopara.2013.09.003}}</ref> Deletion of either of these last two proteins is associated with a reduction in the replication rate in the blood. |
|||
PTEX88 is diffusely located within the blood stage parasites.<ref name="Matz2013"/> In trophozoites, PTEX88 is also localized to previously unrecognized extensions reaching from the parasite surface into the erythrocyte cytoplasm. |
|||
The [[thioredoxin]] 2 protein is part of the multi-protein complex embedded within the parasitophorous vacuolar membrane and is thought to be involved in protein secretion.<ref name=Sharma2011>Sharma A, Sharma A, Dixit S, Sharma A (2011) Structural insights into thioredoxin-2: a component of malaria parasite protein secretion machinery. Sci Rep 1:179</ref> This protein is located in distinct punctate organelles of unknown identity.<ref name="Matz2013"/> |
|||
Within the genome are encoded 11 [[Rab (G-protein)|Rab]] [[GTPase]]s.<ref name=Rached2011>Rached FB, Ndjembo-Ezougou C, Chandran S, Talabani H, Yera H, Dandavate V, Bourdoncle P, Meissner M, Tatu U, Langsley G (2011) Construction of a ''Plasmodium falciparum'' Rab-interactome identifies CK1 and PKA as Rab-effector kinases in malaria parasites. Biol Cell {{doi|10.1111/boc.201100081}}</ref> These proteins are typically involved in vesicle transport. [[Casein kinase]]-1 has been shown to interact with [[RAB5B]] and the catalytic subunit of cAMP-dependent protein kinase A interacts with [[RAB5A]] and [[RAB7]]. |
|||
Several proteins are transported across its plasma membrane, the surrounding parasitophorous vacuole membrane and into its host erythrocyte. Most of these exported proteins contain a host targeting motif. Cleavage is of this motif by the protease plasmepsin V is normally part of this process. This process is not linked to the next component in the export pathway.<ref name=Tarr2012>Tarr SJ, Cryar A, Thalassinos K, Haldar K, Osborne AR (2012) The C-terminal portion of the cleaved HT motif is necessary and sufficient to mediate export of proteins from the malaria parasite into its host cell. Mol Microbiol {{DOI|10.1111/mmi.12133}}</ref> The fifth reside in the host motif is important for the action of plasmepsin V. Mutation of the fourth and fifth positions of this motif, as well as amino acids further downstream, block or affect the efficiency of protein export. |
|||
Surfin 4.1 is a type 1 transmembrane protein expressed on the surface of infected erythrocytes. It is exported to the surface via the endoplasmic reticulum/Golgi apperatus. Its structure varies between strains: in 3D7 there are 19 amino acids after the transmembrane region while FCR3 there are two tryptophan rich domains after the transmembrane domain.<ref name="Zhu2012"/> The transmembrane region is required for the initial movement of the protein to the endoplasmic reticulum. The subsequent sorting step to the parasitophorous vacuole is determined by two independent signals located within the N terminual 50 amino acids. It may form a homodimer during transport. |
|||
[[Sodium]] is actively extruded from the parasite: at least one of the mechanisms involved in this process is PfATP4.<ref name=Spillman2013>Spillman NJ, Allen RJ, McNamara CW, Yeung BK, Winzeler EA, Diagana TT, Kirk K (2013) Na(+) regulation in the malaria parasite ''Plasmodium falciparum'' involves the cation ATPase PfATP4 and is a target of the spiroindolone antimalarials. Cell Host Microbe 13(2) 227-237. {{DOI|10.1016/j.chom.2012.12.006}}</ref> |
|||
A mutation in the mu-chain of the AP2 adaptor complex - a component of the endocytic machinery - has been associated with artemisinin resistance in ''Plasmodium chabaudi''.<ref name=Henriques2013>Henriques G, Martinelli A, Rodrigues L, Modrzynska K, Fawcett R, Houston DR, Borges ST, D Alessandro U, Tinto H, Karema C, Hunt P, Cravo P (2013) Artemisinin resistance in rodent malaria - mutation in the AP2 adaptor mu-chain suggests involvement of endocytosis and membrane protein trafficking. Malar J 12(1) 118</ref> This protein interacts with a cargo recognition protein. |
|||
An essential protein - Cleft-like Protein 1 - has been cloned in ''[[Plasmodium berghei|P. berghei]]''.<ref name=Haase2013>Haase S, Hanssen E, Matthews K, Kalanon M, de Koning-Ward TF (2013) The exported protein PbCP1 localises to cleft-like structures in the rodent malaria parasite ''Plasmodium berghei''" ''PLoS One'' 8(4) e61482. {{DOI|10.1371/journal.pone.0061482}}</ref> This protein has two predicted transmembrane domains in the C-terminal end. It is found in discrete convoluted, vesico-tubular membranous structures in the erythrocyte cytoplasm. |
|||
[[Band 4.1]] is clover leaf shaped protein which interacts with multiple erythrocytic proteins via its three arms known as the N, C and alpha lobes. This protein is involved in maintaining the biconcave shape, elasticity, and mechanical stability of human erythrocytes, and defects in 4.1R are one cause of hereditary erythrocyte [[elliptocytosis]]. The mature parasite-infected erythrocyte surface antigen (MESA) interacts with the C lobe.<ref name=Parish2013>Parish LA, Mai DW, Jones ML, Kitson EL, Rayner JC (2013) A member of the ''Plasmodium falciparum'' PHIST family binds to the erythrocyte cytoskeleton component band 4.1.Malar J 12(1) 160</ref> A secreted protein - PF3D7_0402000 - is localised to the parasitophorous vacuole membrane and interacts with Band 4.1. |
|||
Several of the exported proteins - PfEMP1, PfEMP3, ring associated erythrocyte surface antigen (RESA) and knob associated histidine rich protein (KAHRP) - interact with the preponderant erythrocyte skeleton protein [[spectrin]].<ref name=Weng2013>Weng H, Guo X, Papoin J, Wang J, Coppel R, Mohandas N, An X (2013) Interaction of ''Plasmodium falciparum'' Knob-associated Histidine-rich Protein (KAHRP) with erythrocyte ankyrin R is required for its attachment to the erythrocyte membrane. Biochim Biophys Acta pii: S0005-2736(13)00329-5. {{DOI|10.1016/j.bbamem.2013.09.014}}</ref> KAHRP also binds to [[ankyrin]] R. KAHRP binds to ankyrin via a 79 residue segment: the reciprocal binding site for KAHRP is a subdomain (D3) of the ankyrin R membrane binding domain. KAHRP is normally associated with the host cell membrane. Blocking this interaction with ankyrin R prevents KAHRP movement to the host membrane: instead KAHRP remains diffusely distributed throughout the erythrocyte cytosol. |
|||
===Kinases=== |
|||
Although several [[kinase]]s are known in ''P. falciparum'' (~90 in total<ref name=Talevich2012>Talevich E, Tobin AB, Kannan N, Doerig C (2012) An evolutionary perspective on the kinome of malaria parasites" ''Philos Trans R Soc Lond B Biol Sci'' 367(1602) 2607-2618</ref>) very little is known about them. |
|||
;Cyclin dependent kinases |
|||
A subgroup of cyclin-dependent kinases (CDK) including crk-5 have an activation loop that contains a novel [[Proline]]-[[Threonine]]-x-[[Cytosine]] motif which is absent from all known CDKs outside the Apicomplexa.<ref name=Talevich2011>{{cite journal |author=Talevich E, Mirza A, Kannan N |year=2011 |title=Structural and evolutionary divergence of eukaryotic protein kinases in Apicomplexa |journal=BMC Evol Biol |volume=11 |issue=1 |page=321 |doi=10.1186/1471-2148-11-321}}</ref> |
|||
The protein PFD0975w appears to be homologous with the right open reading frame 2 kinase [[RIO-2]], a kinase involved in [[ribosome]] biogenesis and other cell cycle events.<ref name=Trivedi2012>Trivedi V, Nag S (2012) ''In silico'' characterization of atypical kinase, PFD0975w from ''Plasmodium'' kinome: A suitable target for drug discovery. Chem Biol Drug Des {{doi|10.1111/j.1747-0285.2012.01321.x}}.</ref> This enzyme is unique among the kinases in the genome because along with the kinase domain, it also has a highly conserved N-terminal winged helix domain.<ref name=Chouhan2012>Chouhan DK, Sharon A, Bal C (2012) Molecular and structural insight into plasmodium falciparum RIO2 kinase. J Mol Model</ref> |
|||
The right open reading frame 2 protein kinase may be a potential drug target.<ref name=Nag2012>Nag S, Prasad KM, Bhowmick A, Deshmukh R, Trivedi V (2012) PfRIO-2 kinase is a potential therapeutic target of antimalarial protein kinase inhibitors. Curr Drug Discov Technol</ref> |
|||
;Cyclin dependent like kinases |
|||
Several are [[cyclin]] dependent like kinases (CLK): of these two - the [[Lammer kinase]] homologue PfCLK-1 and PfCLK-2 have been cloned.<ref name="Agarwal2011">Agarwal S, Kern S, Halbert J, Przyborski JM, Baumeister S, Dandekar T, Doerig C, Pradel G. (2011) Two nucleus-localized CDK-like kinases with crucial roles for malaria parasite erythrocytic replication are involved in phosphorylation of splicing factor. J. Cell. Biochem. {{doi|10.1002/jcb.23034}}</ref> CLKs in other eukaryotes are involved in the regulation of [[mRNA]] splicing through phosphorylation of [[serine]]/[[arginine]]-rich proteins. Both are transcribed throughout the asexual blood stages and in gametocytes. PfCLK-1/Lammer possesses two nuclear localization signal sites while PfCLK-2 possesses one of these signal sites upstream of the C-terminal catalytic domains. The two PfCLKs form complexes with proteins with predicted [[nuclease]], [[phosphatase]] or [[helicase]] functions. |
|||
Although the kinases are primarily localized in the parasite nucleus, PfCLK-2 is also present in the cytoplasm. They are important for completion of the asexual replication cycle. Substrates phosphorylated by the PfCLKs include the Sky1p substrate, splicing factor Npl3p, and the plasmodial alternative splicing factor PfASF-1. |
|||
;NIMA kinases |
|||
Within the genome is a family of four protein kinases (Pfnek-1 to -4) that are related to the NIMA ([[never-in-mitosis/Aspergillus]]) family of kinases. The members of this latter family play important roles in [[mitosis]] and [[meiosis]]. Pfnek-1 (PFL1370w) is expressed in asexual parasites and male gametocytes.<ref name=Dorin-Semblat2011>{{cite journal |author=Dorin-Semblat D |year=2011 |title=Plasmodium falciparum'' NIMA-related kinase Pfnek-1: sex specificity and assessment of essentiality for the erythrocytic asexual cycle |journal=Microbiology |volume=157 |issue=10 |pages=2785–2794 |author-separator=, |author2=Schmitt S |author3=Semblat JP |author4=Sicard A |author5=Reininger L |author6=Goldring D |author7=Patterson S |author8=Quashie N |author9=Chakrabarti D |display-authors=9 |doi=10.1099/mic.0.049023-0 |first10=L. |first11=C.}}</ref> It is an essential gene for completion of the asexual cycle. The other three - Pfnek-2 (PFE1290w), -3 (PFL0080c) and -4 (MAL7P1.100) - are expressed predominantly in gametocytes. |
|||
Pfnek-2 is predominantly expressed in gametocytes and is required for DNA replication during meiosis and ookinete development.<ref name=Reininger2009>Reininger L, Tewari R, Fennell C, Holland Z, Goldring D, Ranford-Cartwright L, Billker O, Doerig C (2009) An essential role for the ''Plasmodium'' Nek-2 Nima-related protein kinase in the sexual development of malaria parasites" ''J Biol Chem'' 284(31) 20858-20568</ref> |
|||
The plasmodial mitogen-activated protein kinase kinase Pfnek-3 has both [[serine]]/[[threonine]] and [[tyrosine kinase]] activities.<ref name=Low2011>{{cite journal |author=Low H, Chua CS, Sim TS |year=2011 |title=''Plasmodium falciparum'' possesses a unique dual-specificity serine/threonine and tyrosine kinase, Pfnek3 |journal=Cell Mol Life Sci}}</ref> |
|||
Pfnek-4 is expressed in stage II to V gametocytes and in a subset of asexual stage parasites undergoing schizogony.<ref name=Reininger2012>Reininger L, Garcia M, Tomlins A, Müller S, Doerig C (2012) The ''Plasmodium falciparum'', Nima-related kinase Pfnek-4: a marker for asexual parasites committed to sexual differentiation. Malar J. 11(1) 250.</ref> It is also required for the completion of [[meiosis]] in the ookinete.<ref name=Reininger2009>Reininger L, Tewari R, Fennell C, Holland Z, Goldring D, Ranford-Cartwright L, Billker O, Doerig C (2009) An essential role for the Plasmodium Nek-2 Nima-related protein kinase in the sexual development of malaria parasites" ''J Biol Chem'' 284(31) 20858-20868</ref> |
|||
;Adenylate kinases |
|||
There are at least three [[adenylate kinase]]s (AK) encoded in the genome - PfAK1, PfAK2 and a [[GTP:AMP phosphotransferase]] (PfGAK).<ref name=Ma2012>Ma J, Rahlfs S, Jortzik E, Heiner Schirmer R, Przyborski J, Becker K (2012) Subcellular localization of adenylate kinases in ''Plasmodium falciparum''. FEBS Lett</ref> There are two additional adenylate kinase-like proteins - PfAKLP1 (which is homologous to human AK6) and PfAKLP2. PfAK1, PfAKLP1, and PfAKLP2 are found in the cytosol. PfGAK is located in the mitochondrion. PfAK2 is located at the parasitophorous vacuole membrane and this localization is driven by N-myristoylation. |
|||
[[Adenylate kinase]]s are [[phosphotransferase]]s that catalyze the interconversion of adenine nucleotides. There are at least three adenylate kinases (PfAK1, PfAK2 and GTP:AMP phosphotransferase) encoded in the genome.<ref name=Law2012>Law AW, Lescar J, Hao Q, Kotaka M (2012) Expression, purification, crystallization and preliminary X-ray analysis of ''Plasmodium falciparum'' GTP:AMP phosphotransferase. Acta Crystallogr Sect F Struct Biol Cryst Commun 68(6) 671-674</ref> PfAK1 and PfAK2 both catalyse the conversion of ATP and AMP to two molecules of ADP. PfGAK instead has a preference for GTP and AMP and does not accept ATP as a substrate. |
|||
;Calcium dependent kinases |
|||
The calcium dependent protein kinases (CDPK) are part of a superfamily found in [[plant]]s, [[ciliate]]s and some [[apicomplexa]]. They are not present in [[fungi]] or [[animal]]s. They have three domains: a variable N-terminal region involved in substrate recognition and protein interaction, a kinase catalytic domain and a regulatory domain. The regulatory domain has two subdomains - an autoinhibitory junction domain and a calmodulin like domain. The calmodulin domain has four EF hands. These hands, upon binding calcium, undergo a structural change that moves the junction domain from its autoinhibitory interaction with the substrate binding site of the kinase domain which in turn activates kinase domain catalytic activity. |
|||
Seven CDPKs are present in the genome.<ref name=Lauciello2013>Lauciello L, Kappes B, Scapozza L, Perozzo R (2013) Expression, purification and biochemical characterization of recombinant Ca-dependent protein kinase 2 of the malaria parasite ''Plasmodium falciparum''. Protein Expr Purif pii: S1046-5928(13)00112-5. {{DOI|10.1016/j.pep.2013.06.006}}</ref> The first of these cloned was PfCDPK2 in 1997. |
|||
Calcium dependent protein kinase 1 is expressed in parasite asexual blood and mosquito stages.<ref name=Azevedo2013>Azevedo MF, Sanders PR, Krejany E, Nie CQ, Fu P, Bach LA, Wunderlich G, Crabb BS, Gilson PR (2013) Inhibition of ''Plasmodium falciparum'' CDPK1 by conditional expression of its J-domain demonstrates a key role in schizont development. Biochem J</ref> This protein has a specific auto inhibitory junction region (J). It localises to the parasite plasma membrane of very young intracellular parasites, replicating and invasive forms. It does not appear to be exported into the erythrocyte. Inhibition of this protein results in the arrest of parasite development late in the cell cycle during early schizogony. This protein also appears to have a role in microneme secretion during the process of merozoite invasion of the erythrocyte.<ref name=Bansal2013>Bansal A, Singh S, More KR, Hans D, Nangalia K, Yogavel M, Sharma A, Chitnis CE (2013) Characterization of ''Plasmodium falciparum'' calcium-dependent protein kinase 1 (PfCDPK1) and its role in microneme secretion during erythrocyte invasion. J Biol Chem. 2013 Jul 5;288(27) 19643. {{DOI|10.1074/jbc.A112.411934}}</ref> It also phosphorylates members of the actin-myosin complex.<ref name=Holder2012>Holder AA, Mohd Ridzuan MA, Green JL (2012) Calcium dependent protein kinase 1 and calcium fluxes in the malaria parasite" ''Microbes Infect'' 14(10) 825-30. {{DOI|10.1016/j.micinf.2012.04.006}}</ref> |
|||
Calcium dependent protein kinase 1 autophosphorylates at several site and has an ATP binding site in its N terminal.<ref name=Ahmed2012>Ahmed A, Gaadhe K, Sharma GP, Kumar N, Neculai M, Hui R, Mohanty D, Sharma P (2012) Novel insights into the regulation of malarial calcium-dependent protein kinase 1" ''FASEB J'' 26(8) 3212-21. {{DOI|10.1096/fj.12-203877}}</ref> The protein is myristoylated at its N terminus and is localised to the parasitophorous vacuole and the tubovesicular system of the parasite.<ref name="Möskes2004">Möskes C, Burghaus PA, Wernli B, Sauder U, Dürrenberger M, Kappes B (2004) Export of ''Plasmodium falciparum'' calcium-dependent protein kinase 1 to the parasitophorous vacuole is dependent on three N-terminal membrane anchor motifs" ''Mol Microbiol'' 54(3) 676-691</ref> It is also palmitoylated at its N terminus and has a basic motif located there. |
|||
The homolog of calcium dependent protein kinase 1 (CDPK1) in ''[[Toxoplasma gondii]]'' is calcium dependent protein kinase 3 (TgCDPK3). This protein in ''Toxoplasma'' is localised to the inner membrane and is not an essential gene.<ref name=McCoy2012>McCoy JM, Whitehead L, van Dooren GG, Tonkin CJ (2012) TgCDPK3 regulates calcium-dependent egress of ''Toxoplasma gondii'' from host cells" ''PLoS Pathog'' 8(12) e1003066. {{DOI|10.1371/journal.ppat.1003066}}</ref> It is involved in Ca(2+) ionophore control and host cell egress. The role of this protein in ''Plasmodium'' is not currently known. It is however expressed and localises with proteins at the periphery of the schizonts and merozoites involved in gliding motility<ref name=Kato2008>Kato N, Sakata T, Breton G, Le Roch KG, Nagle A, et al (2008) Gene expression signatures and small-molecule compounds link a protein kinase to ''Plasmodium falciparum'' motility. Nat Chem Biol 4: 347–356</ref> and can can phosphorylate these proteins.<ref name=Green2008>Green JL, Rees-Channer RR, Howell SA, Martin SR, Knuepfer E, et al (2008) The motor complex of ''Plasmodium falciparum'': phosphorylation by a calcium-dependent protein kinase" ''J Biol Chem'' 283: 30980–30989.</ref> Inhibition of CDPK 1 is associated with a block in development at the schizont level.<ref name="Kato2008"/> In ''P bergei'' CDPK1 regulates transcription of stored mRNA during ookinete development in the mosquito midgut.<ref name=Sebastian2012>Sebastian S, Brochet M, Collins MO, Schwach F, Jones ML, Goulding D, Rayner JC, Choudhary JS, Billker O (2012) A ''Plasmodium'' calcium-dependent protein kinase controls zygote development and transmission by translationally activating repressed mRNAs. Cell Host Microbe 12(1) 9-19</ref> |
|||
In ''P. falciparum'' CDPK5 controls parasite egress from host cells.<ref name=Dvorin2012>Dvorin JD, Martyn DC, Patel SD, Grimley JS, Collins CR, (2010) A plant-like kinase in ''Plasmodium falciparum'' regulates parasite egress from erythrocytes" ''Science'' 328: 910–912</ref> In ''P. bergei'' CDPK3 is essential for the ookinete to traverse the mosquito midgut epithelium<ref name=Ishino2006>Ishino T, Orito Y, Chinzei Y, Yuda M (2006) A calcium-dependent protein kinase regulates ''Plasmodium'' ookinete access to the midgut epithelial cell" ''Mol Microbiol'' 59: 1175–1184</ref> and CDPK4 is involved in development of the male gametocyte.<ref name=Billker2004>Billker O, Dechamps S, Tewari R, Wenig G, Franke-Fayard B, et al. (2004) Calcium and a calcium-dependent protein kinase regulate gamete formation and mosquito transmission in a malaria parasite" ''Cell'' 117: 503–514</ref> In ''P. falciparum'' CDPK4 appears to be involved in the exflagelation of the male gametes.<ref name=Ojo2013>Ojo KK, Eastman RT, Vidadala R, Zhang Z, Rivas KL, Choi R, Lutz JD, Reid MC, Fox AM, Hulverson MA, Kennedy M, Isoherranen N, Kim LM, Comess KM, Kempf DJ, Verlinde CL, Su XZ, Kappe S, Maly DJ, Fan E, Van Voorhis WC (2013) A specific inhibitor of PfCDPK4 blocks malaria transmission: Chemical-genetic validation. J Infect Dis </ref> |
|||
;Mitogen activated kinases |
|||
A mitogen activated protein kinase (MAP kinase) gene is located on is located on chromosome 14.<ref name=Lin1996>Lin DT, Goldman ND, Syin C (1996) Stage-specific expression of a Plasmodium falciparum protein related to the eukaryotic mitogen-activated protein kinases. Mol Biochem Parasitol 78(1-2) 67-77</ref> It is predominantly expressed in gametocytes and gametes/zygotes. The protein has 882 amino acid residues and possesses a TDY dual phosphorylation site upstream of the highly conserved VATRWYRAPE sequence within subdomain VIII. Within the carboxyl-terminal segment the protein contains an unusually large and highly charged domain. This region includes two repetitive sequences of either a tetrapeptide or octapeptide motif. |
|||
Two mitogen activated protein kinases are present in the genome.<ref name=Wierk2013>Wierk JK, Langbehn A, Kamper M, Richter S, Burda PC, Heussler VT, Deschermeier C (2013) ''Plasmodium berghei'' MAPK1 displays differential and dynamic subcellular localizations during liver stage development" ''PLoS One'' 8(3) e59755. {{DOI|10.1371/journal.pone.0059755}}</ref> Both genes appear to be transcribed during the liver stages. |
|||
An atypical mitogen activated protein kinase (MAPK) - Pfmap-2 - is known.<ref name=Dorin1999>Dorin D, Alano P, Boccaccio I, Cicéron L, Doerig C, Sulpice R, Parzy D, Doerig C (1999) An atypical mitogen-activated protein kinase (MAPK) homologue expressed in gametocytes of the human malaria parasite ''Plasmodium falciparum''. Identification of a MAPK signature" ''J Biol Chem'' 74(42) 29912-29920</ref> It posses the usual properties of a MAPK - including (i) the ability to undergo autophosphorylation, (ii) the ability to phosphorylate myelin basic protein, a classical MAPK substrate, (iii) the regulation of kinase activity by a MAPK-specific phosphatase and (iv) the ability to be activated by component(s) present in cell extracts. It is expressed in gametocytes. It lacks the conserved threonine-X-tyrosine activation motif usually found in enzymes of this family and instead has a threonine-serine-histidine at the same location. |
|||
; FIKK kinases |
|||
A groups of 20 kinases with a Phe-Ile-Lys-Lys sequence motif appear to be unique to ''P. falciparum''.<ref name=Brandt2013>Brandt GS, Bailey S (2013) Dematin, a human erythrocyte cytoskeletal protein, is a substrate for a recombinant FIKK kinase from ''Plasmodium falciparum''. Mol Biochem Parasitol. 2013 Aug 21. pii: S0166-6851(13)00130-8 {{DOI|10.1016/j.molbiopara.2013.08.003}}</ref> One of these kinases (PfFk4.1, PFD1165w) has been cloned. It autophosphoralates and phosphorlyates [[dematin]], a cytoskeletal protein found at the erythrocyte [[spectrin]]-[[actin]] junction. |
|||
;Other kinases |
|||
The protein kinase CK2, a serine/threonine protein kinase, has one catalytic subunit (PfCK2) and two regulatory ones (PfCK2beta1 and PfCK2beta2).<ref name=Dastidar2012>Dastidar EG, Dayer G, Holland ZM, Dorin-Semblat D, Claes A, Chene A, Sharma A, Hamelin R, Moniatte M, Lopez-Rubio JJ, Scherf A, Doerig C (2012) Involvement of ''Plasmodium falciparum'' protein kinase CK2 in the chromatin assembly pathway. BMC Biol 10(1) 5</ref> This enzyme is found both in the cytoplasm and the nucleus. Substrates include the nucleosome assembly proteins (Naps), histones and two members of the [[Alba]] family. Both of the two regulatory subunits are required for completion of the asexual erythrocytic cycle. |
|||
The cyclic guanine monophosphate dependent protein kinase is essential for the initiation of gametogenesis and for blood stage schizont rupture and may also be involved in ookinete differentiation and motility and liver stage schizont development.<ref name=Hopp2012>Hopp CS, Flueck C, Solyakov L, Tobin A, Baker DA (2012) Spatiotemporal and functional characterisation of the ''Plasmodium falciparum'' cGMP-dependent protein kinase" ''PLoS One'' 7(11) e48206. {{DOI|10.1371/journal.pone.0048206}}</ref> |
|||
[[Pantothenate kinase]], the first enzyme involved in converting [[pantothenate]] to [[coenzyme A]] is present in the genome.<ref name=Spry2013>Spry C, Macuamule C, Lin Z, Virga KG, Lee RE, Strauss E, Saliba KJ (2013) Pantothenamides are potent, on-target inhibitors of ''Plasmodium falciparum'' growth when serum pantetheinase is inactivated" ''PLoS One'' 8(2) e54974 {{DOI|10.1371/journal.pone.0054974}}</ref> It appears to be an essential enzyme. |
|||
A putative O-phosphoseryl-tRNA(Sec) kinase - an enzyme involved in the formation of [[selenocysteine]] tRNA - has been identified in the genome.<ref name=Sherrer2008>Sherrer RL, O'Donoghue P, Söll D (2008) Characterization and evolutionary history of an archaeal kinase involved in selenocysteinyl-tRNA formation" ''Nucleic Acids Res'' 36(4) 1247-59. {{DOI|10.1093/nar/gkm1134}}</ref> A protein kinase C has been identified.<ref name=Sharma2005>Sharma A, Biswas S (2005) Stage-specific cytosolic protein kinase C-like activity in human malarial parasite ''Plasmodium falciparum''. Indian J Biochem Biophys. 2005 Jun;42(3) 145-151</ref> Although predominantly cytosolic it is also present in the membrane faction. Its activation requires Ca<sup>2+</sup>, [[phosphatidyl serine]] and either [[diacylglycerol]] or [[phorbol myristate acetate]]. Its activity is 9 time greater in the trophozoites than in the ring forms. On activiation in the trophozoites the activity in the membrane farction increase significantly. Ita actvity is inhibited in a dose dependent fashion by chloroquine. The inhibition appears to be non compeditive. Chloroquine resistant strains do not show this inhibition of activity. |
|||
A [[tyrosine kinase]] like kinase (PfTKL2) of the IRAK/RLK/Pelle protein family has been identified in the genome.<ref name=Abdi2013>Abdi AI, Carvalho TG, Wilkes JM, Doerig C (2013) A secreted ''Plasmodium falciparum'' kinase reveals a signature motif for classification of tyrosine kinase-like kinases. Microbiology</ref> The gene is expressed in asexual blood stages and in gametocytes. It is also secreted into the culture media. Its function is not presently known. |
|||
Another kinase - PfPK7 - is involved in the rate of asexual growth in erythrocytes, the production merozoites in the schizonts and in the production of oocysts in the mosquito vector.<ref name=Dorin-Semblat2008>Dorin-Semblat D, Sicard A, Doerig C, Ranford-Cartwright L, Doerig C (2008) Disruption of the PfPK7 gene impairs schizogony and sporogony in the human malaria parasite ''Plasmodium falciparum''. Eukaryot Cell 7(2) 279-285</ref> |
|||
===Phosphatases=== |
|||
There are 27 putative protein phosphatases in the genome. These can be classed into groups: phosphoprotein phosphatases, metallo-dependent protein phosphatases, protein tyrosine phosphatases and NLI interacting factor-like phosphatases.<ref name=Wilkes2008>Wilkes JM, Doerig C (2008) The protein-phosphatome of the human malaria parasite ''Plasmodium falciparum''" ''BMC Genomics'' 9: 412</ref> |
|||
A Shewanella-like protein phosphatase that is expressed at all stages of the parasite life cycle but is particularly bundant in asexual blood stages and expressed at all stages of the parasite life cycle is essential for ookinete (zygote) development and microneme formation.<ref name=Patzewitz2013>Patzewitz EM, Guttery DS, Poulin B, Ramakrishnan C, Ferguson DJ, Wall RJ, Brady D, Holder AA, Szöőr B, Tewari R (2013) An ancient protein phosphatase, SHLP1, is critical to microneme development in ''Plasmodium'' ookinetes and parasite transmission. Cell Rep pii: S2211-1247(13)00056-9. {{DOI|10.1016/j.celrep.2013.01.032}}</ref> |
|||
The open reading frame PF3D7_1305500 encodes an atypical MAPK phosphatase. This gene appears to be involved in the regulation of the transition from the pre-S phase to the M phases of asexual intraerythrocytic development.<ref name=Balu2013>Balu B, Campbell C, Sedillo J, Maher S, Singh N, Thomas P, Zhang M, Pance A, Otto TD, Rayner JC, Adams JH (2013) An atypical MAPK phosphatase implicated in regulating transition from pre-S-phase asexual intraerythrocytic development of ''Plasmodium falciparum''. Eukaryot Cell</ref> |
|||
Protein phosphatase 1 is a key enzyme that plays diverse and essential roles in cell biology. Its dephosphorylation activity/specificity is governed by the interaction of its catalytic subunit (PP1c) with regulatory proteins. A homolog of the inhibitor of [[protein phosphatase 1]] has been cloned.<ref name=Freville2011>{{cite journal |author=Freville A, Landrieu I, Garcia-Gimeno MA, Vicogne J, Montbarbon M, Bertin B, Verger A, Kalamou H, Sanz P, Werkmeister E, Pierrot C, Khalife J |year=2011 |title=''Plasmodium falciparum'' inhibitor 3 homolog increases protein phosphatase type 1 activity and is essential for parasitic survival |journal=J Biol Chem}}</ref> This gene is essential for survival and appears to be localised to the nucleus. A conserved 41- [[Lysine]]-[[Valine]]-Valine-[[Arginine]]-[[Tryptophan]]-45 motif is essential for its inhibition activity. Another inhibitor - inhibitor 2 - has also been cloned.<ref name="Frèville2013">Frèville A, Cailliau-Maggio K, Pierrot C, Tellier G, Kalamou H, Lafitte S, Martoriati A, Pierce RJ, Bodart JF, Khalife J (2013) ''Plasmodium falciparum'' encodes a conserved active inhibitor-2 for protein phosphatase type 1: perspectives for novel anti-plasmodial therapy. BMC Biol 11(1) 80</ref> Within the protein are two motifs - 12-[[Lysine]]-[[Threonine]]-[[Isoleucine]]-[[Serine]]-[[Tryptophan]]-16 and 102-[[Histadine]]-[[Tyrosine]]-[[Asparagine]]-[[Glutamine]]-105 - that are critical for its activity. It appears to be an essential gene and seems likely to act at the G2/M point of the cell cycle. |
|||
===Cysteine proteases=== |
|||
A number of cysteine proteases have been identified this organism including four [[falcipain]]s, [[serine repeat antigen]]s (SERA), [[dipeptidyl aminopeptidase]] 1, dipeptidyl aminopeptidase 3 and a [[calpain]] homolog.<ref name=Rosenthal2011>{{cite journal |author=Rosenthal PJ |year=2011 |title=Falcipains and other cysteine proteases of malaria parasites |journal=Adv Exp Med Biol |volume=712 |pages=30–48 |doi=10.1007/978-1-4419-8414-2_3 |pmid=21660657 |series=Advances in Experimental Medicine and Biology |isbn=978-1-4419-8413-5}}</ref> |
|||
;Falcipains |
|||
The falcipains belong to the [[papain]] family of enzymes (clan CA). |
|||
Falcipain-1 appears to be important in the development of the oocysts in the mosquito. The ortholog of this protein in ''[[Plasmodium chabaudi]]'' - chabaupain-1 - is localised preferentially to the apical portion of the ookinete.<ref name=Armada2013>Armada A, Gazarini M, Gonçalves LM, Antunes S, Custódio A, Rodrigues A, Almeida AJ, Silveira H, Rosário VD, Santos-Gomes G, Domingos A (2013) Generation of an antibody that recognizes ''Plasmodium chabaudi'' cysteine protease (chabaupain-1) in both sexual and asexual parasite life cycle and evaluation of chabaupain-1 vaccine potential. Exp Parasitol pii: S0014-4894(13)00167-7. {{DOI|10.1016/j.exppara.2013.06.009}}</ref> |
|||
Falcipain-2 is involved in the hydrolysis of haemoglobin and appears to be a non essential gene. It also promotes host cell rupture by cleaving the skeletal proteins of the erythrocyte membrane. |
|||
Falcipain-3 appears to be an essential gene but its function has yet to be firmly established. |
|||
;Dipeptidyl aminopeptidases |
|||
Dipeptidyl aminopeptidase 1 is found in the digestive vacuole and is also an essential gene. |
|||
Dipeptidyl aminopeptidase 2 is specific to the gametocyte. It does not appear to be an essential gene.<ref name=Tanaka2013>Tanaka TQ, Deu E, Molina-Cruz A, Ashburne MF, Ali O, Suri A, Kortagere S, Bogyo M, Williamson KC (2013) Plasmodium dipeptidyl aminopeptidases as malaria transmission blocking drug targets. Antimicrob Agents Chemother</ref> |
|||
Dipeptidyl aminopeptidase 3 appears to be involved in the release of the merozoites from the erythrocyte. |
|||
;Serine repeat antigens |
|||
The serine repeat antigens are a set of nine proteins in ''P. falciparum'' (two in ''P. gallinaceum'', five in ''P. bergei'', 12 in ''P. vivax'') with a central domain homologous to the papain-like (clan CA, family C1) protease family.<ref name=Ruecker2012>Ruecker A, Shea M, Hackett F, Suarez C, Hirst EM, Milutinovic K, Withers-Martinez C, Blackman MJ (2012) Proteolytic activation of the essential parasitophorous vacuole cysteine protease SERA6 accompanies malaria parasite egress from its host erythrocyte" ''J Biol Chem'' 287(45) 37949-37963. {{DOI|10.1074/jbc.M112.400820}}</ref> They are named after the presence of tandem repeats of serine residues in the protein. |
|||
The only known homolog of the SERA proteases is found in the parasite ''[[Theileria]]''.<ref name=Arisue2007>Arisue N, Hirai M, Arai M, Matsuoka H, Horii T (2007) Phylogeny and evolution of the SERA multigene family in the genus ''Plasmodium''" ''J Mol Evol'' 65:82–91</ref> In a study of 18 species the majority of the SERA genes were found to be clustered between between two conserved genes: a conserved hypothetical protein (HP) gene and the iron-sulfur assembly protein gene (hesB). There is a signal peptide sequence at the N-terminus. The family can be divided into four groups: Groups I to III have a cysteine at the active site and Group IV has a serine residue. Group II and III are sister clades and are more closely related to Group I than to Group IV. With a few exceptions the SERA gene family has a common structure of four exons and three introns. SERA genes of Group I in general have a six exon and five intron structure. In the mammalian malaria parasites, gene duplication occurred only in Group IV SERA genes and was particularly frequent in primate species. The reasons are unknown presently. |
|||
Although most SERAs are cysteine proteases the majority in ''P. falciparum'' (6 of the 9) have a serine residue at the active site. The SERAs possessing a canonical Cys residue at the position of the active site nucleophile in their papain-like central domains are with SERA6, 7 and 8. |
|||
SERA5 and SERA6 are indispensable in blood-stage parasites. SERA 5 (molecular weight 120 kiloDaltons) is abundantly produced at the late trophozoite to schizont stages of parasite development. SERA 6 is produced in much smaller quantities than SERA 5 and appears to be involved with egress from the erythrocyte. SERA 6 is activated by SUB1 - a seine protease that is stored in exonomes. |
|||
SERA8 in ''P. falciparum'' (SERA5 in ''P. berghei'') is expressed exclusively during mosquito stages of the parasite life cycle, where it has been shown to be required for egress of midgut sporozoites from oocysts.<ref name=Aly2005>Aly AS, Matuschewski K (2005) A malarial cysteine protease is necessary for ''Plasmodium'' sporozoite egress from oocysts" ''J Exp Med'' 202: 225–230</ref> |
|||
;Other proteases |
|||
A [[calpain]] homolog has been cloned.<ref name=Soh2013>Soh BY, Song HO, Lee Y, Lee J, Kaewintajuk K, Lee B, Choi YY, Cho JH, Choi S, Park H (2013) Identification of active ''Plasmodium falciparum'' calpain to establish screening system for Pf-calpain-based drug development. Malar J 12(1) 47</ref> Although shorter than other calpains it possesses a typical catalytic triad ([[Cysteine]]-[[Histamine]]-[[Asparagine]]).<ref name=Mitchell2003>Mitchell D, Bell A (2003) PEST sequences in the malaria parasite ''Plasmodium falciparum'': a genomic study. Malar J 2:16</ref> |
|||
A cysteine protease inhibitor - falstatin - is involved in regulating proteolysis during erythrocyte infection.<ref name=Pei2013>Pei Y, Miller JL, Lindner SE, Vaughan AM, Torii M, Kappe SH (2013) ''Plasmodium yoelii'' inhibitor of cysteine proteases is exported to exomembrane structures and interacts with yoelipain-2 during asexual blood stage development. Cell Microbiol {{DOI|10.1111/cmi.12124}}</ref> It is found in vesicles within the asexual blood stage parasite cytoplasm, the parasitophorous vacuole and is exported to dynamic exomembrane structures in the infected erythrocyte. In sporozoites, its is found in the rhoptries and in intracellular vesicles distinct from the micronemes. In the final stages of liver infection it is released into the infected hepatocyte during parasitophorous vacuole membrane breakdown. |
|||
Homologs of these proteins have been identified and cloned in ''[[Plasmodium knowlesi]]''.<ref name=Prasad2012>Prasad R, Atul, Soni A, Puri SK, Sijwali PS (2012) Expression, characterization, and cellular localization of knowpains, papain-like cysteine proteases of the ''Plasmodium knowlesi'' malaria parasite" ''PLoS One'' 7(12) e51619. {{DOI|10.1371/journal.pone.0051619}}</ref> These three knowpains are found in the digestive vacuole, are active at acidic pH and are capable of degrading haemoglobin. Two of them (KP2 and KP3) cleave only if the P2 position is occupied by an [[leucine]] residue. KP4 shows a moderate preference for leucine at the P2 position but is more active if this position is occupied by an [[arginine]] residue. Although found in the digestive vacuole KP4 is also found in the parasite periphery and may play a role in parasite egress from the erythrocyte. |
|||
===Metallopeptidases=== |
|||
There is at least one M1 family aminopeptidase in the genome (PfA-M1). This is a zinc binding metalopeptidase with optimal activity at pH 7.4, and remains at least 40% active between pH 5.8-8.6. It is an alanyl aminopeptidase. Immunofluorescence studies have shown that in trophozoites that it diffusely found in the parasite cytoplasm with accumulations outside the digestive vacuole while in schizonts it is progressively located to a vesicle like pattern ending as a single location in released merozoites. It exists as two major isoforms, a nuclear 120 kDa species and a processed species consisting of a complex of 68 and 35 kDa fragments.<ref name=Ragheb2011>{{cite journal |author=Ragheb D, Dalal S, Bompiani KM, Ray WK, Klemba M |year=2011 |title=Distribution and biochemical properties of an M1-family aminopeptidase in ''Plasmodium falciparum'' indicate a role in vacuolar hemoglobin catabolism |journal=J Biol Chem |doi=10.1074/jbc.M111.225318 |volume=286 |issue=31 |pages=27255–65 |pmid=21659511 |pmc=3149319}}</ref> |
|||
There are at least 2 essential metallopeptidases encoded in the genome - PfA-M1 and Pf-LAP.<ref name=Harbut2011>{{cite journal |author=Harbut MB, Velmourougane G, Dalal S, Reiss G, Whisstock JC, Onder O, Brisson D, McGowan S, Klemba M, Greenbaum DC |year=2011 |title=Bestatin-based chemical biology strategy reveals distinct roles for malaria M1- and M17-family aminopeptidases |journal=Proc Natl Acad Sci USA |doi=10.1073/pnas.1105601108 |volume=108 |issue=34 |pages=E526–34 |pmid=21844374 |pmc=3161592}}</ref> Specific inhibition of PfA-M1 causes swelling of the parasite digestive vacuole and prevented proteolysis of [[haemoglobin]] derived oligopeptides. This inhibition is lethal to the parasite probably by starvation. |
|||
Leucyl aminopeptidase (LAP) is a member of the M17 family. It has a predicted molecular weight of 67.831 kiloDaltons and is localized in the cytosol. It is inhibited by [[bestatin]]. Its inhibition is lethal to parasites early in the life cycle, prior to the onset of haemoglobin degradation suggesting a different role for this enzyme. |
|||
Falcilysin a [[zinc]] [[metalloprotease]] found in the apicoplast.<ref name="Ponpuak2006"/> It is a member of the M16 protease group and has maximal activity at neutral pH. It appears to be an essential gene. Its function in this organelle is not quite clear but it appears to be involved in the degradation of transit peptides. |
|||
M18 AAP is a metallo-aminopeptidase that has a highly restricted specificity for peptides with an N-terminal glutamine or asparagine residue.<ref name=Sivaraman2012>Sivaraman KK, Oellig CA, Huynh K, Atkinson SC, Poreba M, Perugini MA, Trenholme KR, Gardiner DL, Salvesen G, Drag M, Dalton JP, Whisstock JC, McGowan S (2012) X-ray crystal structure and specificity of the Plasmodium falciparum malaria aminopeptidase PfM18AAP. J Mol Biol</ref> |
|||
The gene PFI1625c appears to be a metaloprotease.<ref name=Lhouvum2013>Lhouvum K, Ramakrishnan V, Trivedi V (2013) Insight into structural and biochemical determinants of substrate specificity of PFI1625c: Correlation analysis of protein-peptide molecular models. J Mol Graph Model 43C:21-30. {{DOI|10.1016/j.jmgm.2013.03.008}}</ref> |
|||
===Aspartyl proteases=== |
|||
At least four aspartyl [[protease]]s known as [[plasmepsin]]s are involved in the degradation of [[haemoglobin]] by ''Plasmodium falciparum''.<ref name="Gupta2010">{{cite journal |author=Gupta D, Yedidi RS, Varghese S, Kovari LC, Woster PM |year=2010 |title=Mechanism-based inhibitors of the aspartyl protease plasmepsin II as potential antimalarial agents |journal=J. Med. Chem. |doi=10.1021/jm100233b |volume=53 |issue=10 |pages=4234–47 |pmid=20438064}}</ref> |
|||
The histo-aspartic protease (HAP) has been crystallised.<ref name=Bhaumik2011>Bhaumik P, Xiao H, Hidaka K, Gustchina A, Kiso Y, Yada RY, Wlodawer A (2011) Structural insights into activation and inhibition of histo-aspartic protease (HAP) from ''Plasmodium falciparum''. Biochemistry</ref> This protein has high sequence similarity to [[pepsin]]-like [[aspartic acid|aspartic]] proteases, but one of the two catalytic aspartates, Asp32, is replaced in this enzyme by a [[histidine]] residue. The propeptide interacts with the C-terminal domain of the enzyme, forcing the N- and C- terminal domains apart. This mechanically separates His32 and Asp215 and prevents formation of the mature active site. This mechanism is similar to those of other proplasmepsins. The enzyme has a number of unique features and may be a useful drug target. |
|||
There are at least 10 aspartic proteases encoded within the genome. Plasmepsins I, II, IV and histo-aspartic protease are known to be involved in the digestion of haemoglobin.<ref name=Francis1997>Francis SE, Banerjee R, Goldberg DE (1997) Biosynthesis and maturation of the malaria aspartic hemoglobinases plasmepsins I and II" ''J Biol Chem'' 272(23) 14961-1498</ref> These four enzymes share 50-79% amino acid sequence identity<ref name=Bhaumik2012>Bhaumik P, Gustchina A, Wlodawer A (2012) Structural studies of vacuolar plasmepsins" ''Biochim Biophys Acta'' 1824(1) 207-23. {{DOI|10.1016/j.bbapap.2011.04.008}}</ref> and are located on chromosome 14 (gene identifiers PF14_0076, PF14_0077, PF14_0078, and PF14_0075 respectively).<ref name=Moura2009>Moura PA, Dame JB, Fidock DA (2009) Role of ''Plasmodium falciparum'' digestive vacuole plasmepsins in the specificity and antimalarial mode of action of cysteine and aspartic protease inhibitors" ''Antimicrob Agents Chemother'' 53(12) 4968-4978</ref> Plasmepsins I and II are present in the food vacuole and make the initial cleavages in the hemoglobin molecule. The proplasmepsins I and II are both type II integral membrane proteins that are transported through the secretory pathway before cleavage to the soluble form. This reaction occurs within the food vacuole and the cleavage occurs immediately after a conserved [[Leucine]]-[[Glycine]] dipeptidyl [[sequence motif|motif]].<ref name=Banerjee2003>Banerjee R, Francis SE, Goldberg DE (2003) Food vacuole plasmepsins are processed at a conserved site by an acidic convertase activity in ''Plasmodium falciparum''. Mol Biochem Parasitol 129(2) 157-165</ref> This reaction may be blocked [[calpain]] inhibitors. It appears that plasmepsin II and IV are capable of autoactivation as well as activation each other's inactive form.<ref name=Kim2006>Kim YM, Lee MH, Piao TG, Lee JW, Kim JH, Lee S, Choi KM, Jiang JH, Kim TU, Park H (2006) Prodomain processing of recombinant plasmepsin II and IV, the aspartic proteases of ''Plasmodium falciparum'', is auto- and trans-catalytic. J Biochem 139(2) 189-195</ref> These two proteins are not glycosylated. Plasmepsin I is synthesized and processed to the mature form soon after the parasite invades the red blood cell, while plasmepsin II synthesis is delayed until later in development. |
|||
;Intramembrane proteases |
|||
Plasmepsin V, an integral membrane protein, is located within the endoplasmic reticulum but not in the Golgi apperatus.<ref name=Klemba2005>Klemba M, Goldberg DE (2005) Characterization of plasmepsin V, a membrane-bound aspartic protease homolog in the endoplasmic reticulum of ''Plasmodium falciparum''. Mol Biochem Parasitol 143(2) 183-191</ref> The gene is expressed over the course of asexual intraerythrocytic development. The amount of the protein in the parasite is lowest in the ring stage and increases steadily through schizogony. It appears to be involved in the export of proteins to the erythrocyte. Plasmepsin V cleaves N-terminal sequences from RIFIN, STEVOR and RESA multigene families but does not cleave the N-terminal sequence of erythrocyte membrane protein 1, skeleton binding protein (SBP-1) or REX-2.<ref name=Boddey2013>Boddey JA, Carvalho TG, Hodder AN, Sargeant TJ, Sleebs BE, Marapana D, Lopaticki S, Nebl T, Cowman AF (2013) Role of Plasmepsin V in export of diverse protein families from the ''Plasmodium falciparum'' exportome. Traffic {{DOI|10.1111/tra.12053}}</ref> |
|||
A [[presenilin]]-like [[signal peptide peptidase]] is known to be present in the [[endoplasmic reticulum]].<ref name=Marapana2012>Marapana DS, Wilson DW, Zuccala ES, Dekiwadia CD, Beeson JG, Ralph SA, Baum J (2012) Malaria parasite signal peptide peptidase is an ER-resident protease required for growth but not invasion. Traffic {{DOI|10.1111/j.1600-0854.2012.01402.x}}</ref> |
|||
===Serine proteases=== |
|||
A [[SUMO protein|SUMO]] specific protease PfSENP1 (PFL1635w) has been identified in the genome but its importance if any is not known<ref name=Ponder2011>{{cite journal |author=Ponder EL |year=2011 |title=Functional characterization of a SUMO deconjugating protease of Plasmodium falciparum using newly identified small molecule inhibitors |journal=Chem Biol. |volume=18 |issue=6 |pages=711–721 |doi=10.1016/j.chembiol.2011.04.010 |pmid=21700207 |pmc=3131532 |author-separator=, |author2=Albrow VE |author3=Leader BA |author4=Békés M |author5=Mikolajczyk J |author6=Fonović UP |author7=Shen A |author8=Drag M |author9=Xiao J |display-authors=9 |first10=Edgar |first11=Amy J.}}</ref> |
|||
There are at least 3 [[subtilisin]] like proteases encoded in the genome.<ref name=Alam2012>Alam A, Bhatnagar RK, Chauhan VS (2012) Expression and characterization of catalytic domain of ''Plasmodium falciparum'' subtilisin-like protease 3. Mol Biochem Parasitol</ref> These are serine proteases. |
|||
SUB1 has multiple roles in the parasite's life cycle: in the blood stages it is involved in the egress of merozoites from the infected erythrocytes and also in priming merozoites for subsequent erythrocyte invasion.<ref name=Tawk2013>Tawk L, Lacroix C, Gueirard P, Kent R, Gorgette O, Thiberge S, Mercereau-Puijalon O, Ménard R, Barale JC (2013) A key role for ''Plasmodium'' subtilisin-like SUB1 in egress of malaria parasites from host hepatocytes. J Biol Chem</ref> It also appears to be involved in escaping from the parasitophorous vacuole membrane in the hepatocytes. Within the merozoites SUB1 is stored in exonemes<ref name=Yeoh2007>Yeoh S, O'Donnell RA, Koussis K, Dluzewski AR, Ansell KH, Osborne SA, Hackett F, Withers-Martinez C, Mitchell GH, Bannister LH, Bryans JS, Kettleborough CA, Blackman MJ (2007) Subcellular discharge of a serine protease mediates release of invasive malaria parasites from host erythrocytes" ''Cell'' 131(6) 1072-1083</ref> and released in response to a rise in calcium.<ref name=Agarwal2013>Agarwal S, Singh MK, Garg S, Chitnis CE, Singh S (2013) Ca(2+)-mediated exocytosis of subtilisin-like protease 1: a key step in egress of ''Plasmodium falciparum'' merozoites. Cell Microbiol 15(6) 910-921. {{DOI|10.1111/cmi.12086}}</ref> Blocking this release results in failure to process serine repeat antigen 5 (PfSERA5) and parasitophorous vacuolar membrane rupture and merozoite egress from the hepatocytes. SUB1 also acts on merozoite surface protein 1 (MSP1), MSP6 and MSP7 on the merozoite surface.<ref name=Koussis2009>Koussis K, Withers-Martinez C, Yeoh S, Child M, Hackett F, Knuepfer E, Juliano L, Woehlbier U, Bujard H, Blackman MJ (2009) A multifunctional serine protease primes the malaria parasite for red blood cell invasion" ''EMBO J'' 28(6) 725-735. {{DOI|10.1038/emboj.2009.22}}</ref> Blocking the action of SUB1 on these proteins reduces the ability of the merozoite to invade erythrocytes. SUB1 also acts on serine repeat antigen 6 (SERA 6) - a cysteine protease found in the intraerythrocytic parasitophorous vacuole.<ref name=Ruecker2012>Ruecker A, Shea M, Hackett F, Suarez C, Hirst EM, Milutinovic K, Withers-Martinez C, Blackman MJ (2012) Proteolytic activation of the essential parasitophorous vacuole cysteine protease SERA6 accompanies malaria parasite egress from its host erythrocyte" ''J Biol Chem'' 287(45) 37949-63. {{DOI|10.1074/jbc.M112.400820}}</ref> The action of SUB1 activates the proteolytic function of SERA 6. |
|||
In the merozoites SUB2 has been implicated in shedding of adhesins at a juxtamembrane position.<ref name=Harris2005>Harris PK, Yeoh S, Dluzewski AR, O'Donnell RA, Withers-Martinez C, Hackett F, Bannister LH, Mitchell GH, Blackman MJ (2005) Molecular identification of a malaria merozoite surface sheddase" ''PLoS Pathog'' 1(3) 241-251</ref> PfSUB2 is located on the surface of the parasite.<ref name=Child2013>Child MA, Harris PK, Collins CR, Withers-Martinez C, Yeoh S, Blackman MJ (2013) Molecular determinants for subcellular trafficking of the malarial sheddase PfSUB2. Traffic {{DOI|10.1111/tra.12092}}</ref> Posttranslationally it is transported first to the micronemes and then to the surface. Its usual promoter is required for its final location. Its transmembrane domain is required for transport to the microneme and the cytoplasmic domain is required for its surface localization. |
|||
Another protease - PfSUB3 - is expressed at late asexual blood stages. This enzyme is known to act on [[profilin]] (PfPRF).<ref name=Alam2013>Alam A, Bhatnagar RK, Relan U, Mukherjee P, Chauhan VS (2013) Proteolytic activity of ''Plasmodium falciparum'' subtilisin-like protease 3 on parasite profilin, a multifunctional protein. Mol Biochem Parasitol pii: S0166-6851(13)00137-0. {{DOI|10.1016/j.molbiopara.2013.09.006}}</ref> |
|||
;Intramembrane proteases |
|||
At least two [[rhomboid protease]]s (ROM1 and ROM4) are present in the genome.<ref name="Ejigiri2012"/> They are able to act on a number of substrates including TRAP, CTRP, MTRAP, PFF0800c, EBA-175, BAEBL, JESEBL, MAEBL, AMA1, Rh1, Rh2a, Rh2b and Rh4 but not PTRAMP.<ref name=Baker2006>Baker RP, Wijetilaka R, Urban S (2006) Two ''Plasmodium'' rhomboid proteases preferentially cleave different adhesins implicated in all invasive stages of malaria" ''PLoS Pathog'' 2(10) e113</ref> |
|||
The protease rhomboid-1 (ROM1: PF11_0150) is located within a single, thread-like structure on one side of the merozoites that appears to be in close proximity to the subpellicular microtubules. This organelle has been named the mononeme.<ref name=Singh2007>Singh S, Plassmeyer M, Gaur D, Miller LH (2007) Mononeme: a new secretory organelle in ''Plasmodium falciparum'' merozoites identified by localization of rhomboid-1 protease" ''Proc Natl Acad Sci USA'' 104(50) 20043-20048</ref> The mononeme first appears in the schizonts. On the release of merozoites from schizonts, ROM1 moves from a lateral asymmetric localization to the merozoite apical pole and the posterior pole. It can cleave the apical membrane antigen 1.<ref name=Baker2006>Baker RP, Wijetilaka R, Urban S (2006) Two ''Plasmodium'' rhomboid proteases preferentially cleave different adhesins implicated in all invasive stages of malaria" ''PLoS Pathog'' 2(10) e113</ref> |
|||
ROM4 (PFE0340c) appears to be involved in the gliding motility of the sporozoite. |
|||
===Actin=== |
|||
Within the genome are encoded two forms of the protein [[actin]] - I and II. The first form (I) is present in significantly greater quantities. Actin II appears to be essential for the process of exflagellation.<ref name=Deligianni2011>Deligianni E, Morgan RN, Bertuccini L, Kooij TW, Laforge A, Nahar C, Poulakakis N, Schüler H, Louis C, Matuschewski K, Siden-Kiamos I (2011) Critical role for a stage-specific actin in male exflagellation of the malaria parasite. Cell Microbiol {{doi|10.1111/j.1462-5822.2011.01652.x}}</ref> Deletion of this gene results in viable asexual stages. During the formation of the male gametes actin I is found initially in both the nucleus and the cytoplasm. After activation it is found only in the cytoplasm. In actin II deletion mutants actin I remains in both the nucleus and the cytoplasm after activation. Morphologically in the actin II mutants male gametocyte DNA was replicates normally and axonemes are assembled but egress from the host cell is inhibited and axoneme motility is abolished. |
|||
Two proteins ''P. falciparum'' actin-depolymerizing factor 1 (PfADF1) and ''P. falciparum'' actin-depolymerizing factor 2 (PfADF2) are involved in the polymerisation of actin.<ref name=Wong2011>{{cite journal |author=Wong W, Skau CT, Marapana DS, Hanssen E, Taylor NL, Riglar DT, Zuccala ES, Angrisano F, Lewis H, Catimel B, Clarke OB, Kershaw NJ, Perugini MA, Kovar DR, Gulbis JM, Baum J |year=2011 |title=Minimal requirements for actin filament disassembly revealed by structural analysis of malaria parasite actin-depolymerizing factor 1 |journal=Proc Natl Acad Sci USA |doi=10.1073/pnas.1018927108 |volume=108 |issue=24 |pages=9869–74 |pmid=21628589 |pmc=3116436}}</ref> PfADF1 has ben crystallised and despite having significant differences from other proteins with similar function it is capable of severing actin filaments. PfADF2, like canonical ADF proteins but unlike ADF1, binds to both globular and filamentous actin, severing the filaments and inducing nucleotide exchange on the actin monomer.<ref name=Singh2011>{{cite journal |author=Singh BK, Sattler JM, Chatterjee M, Huttu J, Schüler H, Kursula I |year=2011 |title=Crystal structures explain functional differences in the two actin depolymerization factors of the malaria parasite |journal=J Biol Chem |volume=286 |issue=32 |pages=28256–28264 |doi=10.1074/jbc.M111.211730 |pmid=21832095 |pmc=3151070}}</ref> The crystal structure of PfADF1 shows major differences from the ADF consensus, explaining the lack of F-actin binding. PfADF2 structurally resembles the canonical members of the ADF/cofilin family. |
|||
The actins found In ''Plasmodium'' and in ''[[Toxoplasma]]'' are divergent both in sequence and function and only form short, unstable filaments in contrast to the stability of conventional actin filaments.<ref name=Skillman2011>{{cite journal |author=Skillman KM, Diraviyam K, Khan A, Tang K, Sept D, Sibley LD |year=2011 |title=Evolutionarily divergent, unstable filamentous actin is essential for gliding motility in apicomplexan parasites |journal=PLoS Pathog |volume=7 |issue=10 |page=e1002280 |doi=10.1371/journal.ppat.1002280 |editor1-last=Striepen |editor1-first=Boris}}</ref> This inherent instability of parasite's actin filaments is a critical adaptation for their gliding motility. |
|||
Actin is involved in the expression of the ''var'' genes.<ref name=Zhang2011>Zhang Q, Huang Y, Zhang Y, Fang X, Claes A, Duchateau M, Namane A, Lopez-Rubio JJ, Pan W, Scherf A (2011) A critical role of perinuclear filamentous actin in spatial repositioning and mutually exclusive expression of virulence genes in malaria parasites. Cell Host Microbe 10(5) 451-463</ref> The ''var'' introns interact with an 18 base pair nuclear protein binding element which recruits actin and repositions the ''var'' DNA from a transcriptionally repressive to a transcriptionally active perinuclear compartment. |
|||
The presence of actin microfilaments has been demonstrated in the ookinete in the [[pellicle (biology)|pellicle]] and in the apices.<ref name=Angrisano2011>{{cite journal |author=Angrisano F, Delves MJ, Sturm A, Mollard V, McFadden GI, Sinden RE, Baum J |year=2011 |title=A GFP-actin reporter line to explore microfilament dynamics across the malaria parasite lifecycle |journal=Mol Biochem Parasitol}}</ref> |
|||
There are two [[formin]] genes encoded in the genome.<ref name=Ignatev2012>{{cite journal |last1=Ignatev |first1=A |last2=Bhargav |first2=SP |last3=Vahokoski |first3=J |last4=Kursula |first4=P |last5=Kursula |first5=I |year=2012 |title=The lasso segment is required for functional dimerization of the ''Plasmodium'' formin 1 FH2 domain |journal=PLoS ONE |volume=7 |issue=3 |page=e33586 |doi=10.1371/journal.pone.0033586 |editor1-last=Frischknecht |editor1-first=Friedrich}}</ref> These associate with and nucleate both mammalian and ''Plasmodium'' actin filaments. Another gene [[profilin]] - also encoded in the genome but only as a single copy - sequesters actin monomers preventing their polymerisation. |
|||
[[Aldolase]], an actin binding protein, is involved in the moving junction that forms during the invasion of the erythrocyte.<ref name=Uchime2012>Uchime O, Herrera R, Reiter K, Kotova S, Shimp RL Jr, Miura K, Jones D, Lebowitz J, Ambroggio X, Hurt DE, Jin AJ, Long C, Miller LH, Narum DL (2012) Analysis of the conformation and function of the ''Plasmodium falciparum'' merozoite proteins MTRAP and PTRAMP. Eukaryot Cell</ref> |
|||
Several hemoglobinopathies that protect carriers from severe malaria may do so by interfering with host actin reorganization.<ref name=Cyrklaff2012>Cyrklaff M, Sanchez CP, Frischknecht F, Lanzer M (2012) Host actin remodeling and protection from malaria by hemoglobinopathies. Trends Parasitol pii: S1471-4922(12)00143-2 {{DOI|10.1016/j.pt.2012.08.003}}</ref> |
|||
The cyclase associated proteins are among the most highly conserved regulators of actin dynamics. They catalyze nucleotide exchange on actin monomers from ADP to ATP and recycle actin monomers from ADF/cofilin for new rounds of filament assembly. The ''Plasmodium falciparum'' cyclase associated protein is entirely composed of β-sheet domains and efficiently promotes nucleotide exchange on actin monomers.<ref name=Makkonen2012>Makkonen M, Bertling E, Chebotareva N, Baum J, Lappalainen P (2012) Mammalian and malaria parasite cyclase-associated proteins catalyze nucleotide exchange on G-actin Through a conserved mechanism. J Biol Chem</ref> |
|||
===Ubiquitin=== |
|||
The addition of the small protein [[ubiquitin]] to other proteins as part of post translational processing is widespread in most eukaryotes. This is also the case with ''P. falciparum'' where this process occurs at all stages of the asexual life cycle.<ref>{{cite journal |author=Ponts N, Saraf A, Chung DW, Harris A, Prudhomme J, Washburn MP, Florens L, Le Roch KG |year=2011 |title=Unraveling the human malaria parasite's ubiquitome |journal=J Biol Chem |doi=10.1074/jbc.M111.238790 |volume=286 |issue=46 |pages=40320–30 |pmid=21930698 |pmc=3220526}}</ref> Ubiquitylation involves the covalent attachment of a ubiquitin moiety to [[lysine]] residues of protein substrates via the hierarchical intervention of an E1 ubiquitin activating enzyme, an E2 ubiquitin conjugating enzyme, and an E3 ubiquitin ligase that is usually involved in specific substrate recognition. This process is also known as sumoylation (small ubiquitin related modifiers). |
|||
Ubiquitylation is involved in removing misfolded proteins from the [[endoplasmic reticulum]] - a process known as [[Endoplasmic-reticulum-associated protein degradation]]. This process is a prerequisite for subsequent retro-translocation to the cytosol and destruction by the [[proteasome|26S proteasome]]. Aberrant proteins are recognized by endoplasmic reticulum luminal chaperone proteins and protein disulfide isomerases to help discriminate properly folded proteins from misfolded proteins. Misfolded proteins are shuttled to the DER1 translocon complex which forms a hydrophobic pore to allow the retro-translocation of proteins through the endoplasmic reticulum membrane. Several components of the system are known to be present in the parasite: HRD1 (E3 ubiquitin ligase), UBC (E2 ubiquitin conjugating enzyme) and UBA1 (E1 ubiquitin activating enzyme).<ref name="Chung2012"/> HRD1 localizes to the endoplasmic reticulum membranes, while UBC and UBA1 localize to the cytosol. HRD1 interacts with membrane bound proteins needed for retro-translocation and helps form the hydrophobic pore complex. Another member of this pathway is the [[signal peptide peptidase]]. |
|||
The enzymatic mechanism is reasonably well understood. The E1 enzyme adenylates ubiquitin at its C-terminus, creating a mixed anhydride: this process requires ATP. The sulfhydryl group of the E1 active site [[cysteine]] then attacks the anhydride: thus results in the formation of a high energy thio-ester linking ubiquitin to E1. Ubiquitin is then passed to the active site cysteine of the E2 enzyme. Finally, with the aid of an E3 ligase, ubiquitin is transferred from E2 and covalently attached to the ε-amino group of a [[lysine]] in the target protein. |
|||
The genes PFL1245w is the E1 ubiquitin activating enzyme, PFL0190w is the E2 ubiquitin conjugating enzyme and PF14_0215 is the E3 ubiquitin ligase. PFL1245w (E1) contains a ubiquitin activating enzyme active site, two ubiquitin like activating enzyme catalytic domains, two ThiF repeats and a catalytic cysteine at the N-terminal end. PFL0190w (E2) is 147 amino acid residues in length and contains an ubiquitin conjugating enzyme domain takes up almost its whole length. PF14_0215 (E3) has multiple transmembrane domains, an E3 RING zinc finger (zf-C3HC4) domain on its C-terminal half and a predicted [[signal peptide]] consistent with endoplasmic reticulum targeting. The presence of four transmembrane domains is compatible with a pore forming ability and to be able to participate in the recognition and translocation of misfolded proteins across the endoplasmic reticulum membrane. |
|||
The addition of ubiquitin to a protein frequently precceds its digestion in the [[proteasome]]. The proteasome mediates the nonlysosomal degradation of cytosolic proteins in eukaryotic cells. It is a large complex consisting of two multisubunit structures, the 20S and 19S (PA700) or P28 complexes which combine to form the 26S particles. The proteasome subunit 4 ATPase has been cloned from ''P. falciparum''.<ref name=Certad1999>Certad G, Abrahem A, Georges E (1999) Cloning and partial characterization of the proteasome S4 ATPase from ''Plasmodium falciparum''. Exp Parasitol 93(3) 123-131</ref> The protein is similar to those found in eukaryotes. It lacks introns. |
|||
The E1 and E2 enzymes interact to recognize and modify RanGAP1.<ref name=Reiter2013>Reiter K, Mukhopadhyay D, Zhang H, Boucher LE, Kumar N, Bosch J, Matunis MJ (2013) Identification of biochemically distinct properties of the SUMO conjugation pathway in ''Plasmodium falciparum''. J Biol Chem</ref> Sumoylation actvity peaks during mid stages of the intra-erythrocyte developmental cycle. |
|||
A mutation in a deubiquitinating enzyme in ''Plasmodium chabaudi'' has been reported to cause both artesunate and chloroquine resistance.<ref name=Hunt2007>Hunt P, Afonso A, Creasey A, Culleton R, Sidhu AB, Logan J, Valderramos SG, McNae I, Cheesman S, do Rosario V, Carter R, Fidock DA, Cravo P (2007) Gene encoding a deubiquitinating enzyme is mutated in artesunate- and chloroquine-resistant rodent malaria parasites" ''Mol Microbiol'' 65(1) 27-40</ref> |
|||
It appears that ubiquitylation may be a mechanistic requirement of apicoplast protein import independent of the proteasomal degradation pathway.<ref name=Agrawal2013>Agrawal S, Chung DW, Ponts N, van Dooren GG, Prudhomme J, Brooks CF, Rodrigues EM, Tan JC, Ferdig MT, Striepen B, Le Roch KG (20130 An apicoplast localized ubiquitylation system is required for the import of nuclear-encoded plastid roteins" ''PLoS Pathog'' 9(6) e1003426. {{DOI|10.1371/journal.ppat.1003426}}</ref> |
|||
===Heat shock proteins=== |
|||
A number of [[heat shock protein]]s 40 (hsp40) have been predicted from the sequenced genome. Only one is predicted to be a cytosolic canonical Hsp40 capable of interacting with the major cytosolic Hsp70 an interaction that has been confirmed experimentally.<ref name="Botha2010">{{cite journal |author=Botha M, Chiang AN, Needham PG, Stephens LL, Hoppe HC, Külzer S, Przyborski JM, Lingelbach K, Wipf P, Brodsky JL, Shonhai A, Blatch GL |year=2010 |title=''Plasmodium falciparum'' encodes a single cytosolic type I Hsp40 that functionally interacts with Hsp70 and is upregulated by heat shock |journal=Cell Stress Chaperones}}</ref> |
|||
Heat shock protein 20 has been shown to have a critical role in sporozoite motility.<ref name=Montagna2011>Montagna GN, Buscaglia CA, Munter S, Goosmann C, Frischknecht F, Brinkmann V, Matuschewski K (2011) Critical role for heat shock protein 20 (HSP20) in migration of malarial sporozoites. J Biol Chem</ref> This role appears to be via substrate adhesion. |
|||
PfGECO is a type IV heat shock protein 40 expressed in gametocyte stages I to IV and is exported to the erythrocyte cytoplasm.<ref name=Morahan2011>{{cite journal |author=Morahan BJ, Strobel C, Hasan U, Czesny B, Mantel PY, Marti M, Eksi S, Williamson KC |year=2011 |title=Functional analysis of the exported type IV HSP40 protein PfGECO in ''P. falciparum'' gametocytes |journal=[[Eukaryotic Cell]] |doi=10.1128/EC.05155-11 |volume=10 |issue=11 |pages=1492}}</ref> This gene appears to be non essential. |
|||
A Hsp40 class of chaperone (PFB0090c; PF3D7_0201800; KAHsp40) is located in a chromosomal cluster together with knob components KAHRP and PfEMP3.<ref name=Acharya2012>Acharya P, Chaubey S, Grover M, Tatu U (2012) An exported heat shock protein 40 associates with pathogenesis-related knobs in ''Plasmodium falciparum'' infected erythrocytes" ''PLoS One'' 7(9) e44605</ref> This protein has a PEXEL motif required for transport to the erythrocyte compartment. It occurs in punctuate spots in the erythrocyte periphery, distinctly from Maurer's clefts. These structures may be knobs particularly since it is found in a complex the known knob proteins KAHRP, PfEMP3 and Hsp101. |
|||
There are 6 HSP70 proteins in the genome.<ref name=Hatherley2013>Hatherley R, Blatch GL, Bishop OT (2013) ''Plasmodium falciparum'' Hsp70-x: a heat shock protein at the host-parasite interface. J Biomol Struct Dyn</ref> Five of these proteins are found in other species: the exception being PfHsp70-x. |
|||
Heat shock proteins Hsp70 and Hsp90 are both expressed in ''P. falciparum''. They are linked by an essential adaptor protein known as the Hsp70-Hsp90 organising protein (Hop). This protein co-localises with PfHsp70 and PfHsp90 at the trophozoite stage and forms a complex with them.<ref name=Gitau2011>Gitau GW, Mandal P, Blatch GL, Przyborski J, Shonhai A (2011) Characterisation of the ''Plasmodium falciparum'' Hsp70-Hsp90 organising protein (PfHop). Cell Stress Chaperones</ref> |
|||
A heat shock protein PfHsp70-x with endoplasmic reticulum signal peptide has been identified.<ref name=Grover2013>Grover M, Chaubey S, Ranade S, Tatu U (2013) Identification of an exported heat shock protein 70 in ''Plasmodium falciparum''. Parasite 20:2</ref> It is maximally expressed at the schizont stage of intra erythrocytic life cycle. Although the majority of the protein localizes to the parasitophorous vacuole, some of it gets exported to the erythrocyte compartment where it associates with Maurer's clefts. It lacks an endoplasmic reticumlum signal and interacts with at least some of the HSP40 proteins. |
|||
The protein [[Aha1]] interacts with HSP90.<ref name=Chua2011>{{cite journal |author=Chua CS, Low H, Lehming N, Sim TS |year=2011 |title=Molecular analysis of Plasmodium falciparum co-chaperone Aha1 supports its interaction with and regulation of Hsp90 in the malaria parasite |journal=Int J Biochem Cell Biol}}</ref> |
|||
The heat shock protein 110 (Hsp110) is an essential protein in ''P. falciparum''.<ref name=Muralidharan2012>Muralidharan V, Oksman A, Pal P, Lindquist S, Goldberg DE (2012) ''Plasmodium falciparum'' heat shock protein 110 stabilizes the asparagine repeat-rich parasite proteome during malarial fevers. Nat Commun 3:1310. {{DOI|10.1038/ncomms2306}}</ref> Although its usual function is still under investigation it is known to prevent the aggregation of [[asparagine]] repeat rich proteins within the parasite. These repeat rich proteins tend to form aggregates particularly at the elevated temperatures that occur with malaria. |
|||
A 20 kiloDalton protein (cpn20) that acts as a heat shock protein and is localised in the apicoplast has been cloned.<ref name=Vitlin2013>Vitlin Gruber A, Nisemblat S, Zizelski G, Parnas A, Dzikowski R, Azem A, Weiss C (2013) ''P. falciparum'' cpn20 Is a bona fide co-chaperonin that can replace GroES in ''E. coli''" ''PLoS One'' 8(1) e53909. {{DOI|10.1371/journal.pone.0053909}}</ref> |
|||
===Redox balance=== |
|||
The infected host cell is under considerable oxidative stress. Normal erythrocytes have a ratio of reduced (GSH) and oxidized glutathione (GSSG) of 321.6 while the GSH/GSSG ratio in infected cells is 26.7.<ref name=Atamna1997>Atamna H, Ginsburg H (1997) The malaria parasite supplies glutathione to its host cell - investigation of glutathione transport and metabolism in human erythrocytes infected with ''Plasmodium falciparum''" ''Eur J Biochem'' 250(3) 670-679</ref> The ratio in the parasite is 284.5. Efflux of GSSG from the intact infected cell is more than 60-fold higher than the rate observed in normal erythrocytes. This export process is mediated by permeability pathways that the parasite induces in the erythrocyte's membrane. Exogenous gamma-glutamylcysteine is not converted into GSH in the infected erythrocyte suggesting that the erythrocytes' own GSH synthetase may not be functional. This may be due to the lower levels of [[magnesium]] (Mg<sup>2+</sup>) in the infected erythrocyte (0.5 milliMolar) compared to the normal erythrocytes (1.5-3 mM). The lower level of results in cessation of gamma-glutamylcysteine synthesis and of GSH synthesis in the infected erythrocyte. The parasite maintains a level of 4 mM magnesium. The parasite membrane is impermeable to both gamma-glutamylcysteine and GSH. |
|||
The proteins [[gamma-glutamylcysteine synthetase]] and [[glutathione synthetase]] which are involved in the synthesis of [[glutathione]] appear to be essential genes in ''P. falciparum''.<ref name=Patzewitz2011>Patzewitz EM, Wong EH, Müller S (2011) Dissecting the role of glutathione biosynthesis in ''Plasmodium falciparum''. Mol Microbiol {{doi|10.1111/j.1365-2958.2011.07933.x}}</ref> Inhibition of the glutathione biosynthesis by the parasite is lethal. Its levels appear to be tightly regulated. The enzyme [[glutathione reductase]] is highly specific for its substrate [[glutathione disulfide]].<ref name=Jortzik2012>Jortzik E, Becker K (2012) Thioredoxin and glutathione systems in ''Plasmodium falciparum''. Int J Med Microbiol</ref> |
|||
Glutathione export from parasitized cells is inhibited partially by both the compound MK571 and by [[furosemide]].<ref name=Barrand2012>Barrand MA, Winterberg M, Ng F, Nguyen M, Kirk K, Hladky SB (2012) Glutathione export from human erythrocytes and ''Plasmodium falciparum'' malaria parasites. Biochem J</ref> These agents are inhibitors of the 'new permeability pathways' induced by the parasite in the host erythrocyte membrane. |
|||
A possible connextion between the redox system and chloroquine resistance has been described.<ref name=Patzewitz2012>Patzewitz EM, Salcedo-Sora JE, Wong EH, Sethia S, Stocks PA, Maughan SC, Murray JA, Krishna S, Bray PG, Ward SA, Müller S (2012) Glutathione transport: A new role for PfCRT in chloroquine resistance. Antioxid Redox Signal</ref> Strains carrying the mutant chloroquine resistant transport gene have lower levels of glutathione and are more sensitive to inhibition of glutathione synthesis. The mutant chloroquine resistance transport gene selectively transports glutathione into the digestive vacuole. Also in the mutant strains the haemozoin levels are lower and there is reduced chloroquine binding by haemozoin. It seems likely that the increased transport of glutathione into the digestive vacuole may alter both the synthesis of haemozoin within the digestive vacuole and the binding of chloroquine to haemozoin within the digestive vacuole. |
|||
The thioredoxin system like the glutathione system is responsible for maintaining the [[redox]] balance in the cell. The [[thioredoxin reductase]] reduces [[thioredoxin]] and a number of other low molecular weight compounds.<ref name="Jortzik2012"/> The other members of this system include five [[peroxiredoxin]]s differentially located in the cytosol, apicoplast, mitochondria and nucleus with partially overlapping substrate preferences. It also includes members of the thioredoxin superfamily with three thioredoxins, two thioredoxin-like proteins, a dithiol and three monocysteine [[glutaredoxin]]s and a redox-active [[plasmoredoxin]] being encoded in the genome. |
|||
Within the cytoplasm two peroxiredoxins - T peroxiredoxin-1 (TPx-1) and 1-Cys peroxiredoxin (1-Cys Prx) - are produced at differing points in the life cycle.<ref name=Kimura2012>Kimura R, Komaki-Yasuda K, Kawazu SI, Kano S (2012) 2-Cys peroxiredoxin of ''Plasmodium falciparum'' is involved in resistance to heat stress of the parasite. Parasitol Int pii: S1383-5769(12)00157-2. {{DOI|10.1016/j.parint.2012.11.005}}</ref> Disruption of the T peroxiredoxin-1 enzymes renders the parasite hypersensitive to heat stress. This does not occur with knock out mutants of 1-Cys peroxiredoxin suggesting that these enzymes have different roles in the life cycle. |
|||
Investigation of the liver stages of these enzymes in ''[[Plasmodium berghei]]'' has shown that both TPx-1 and 1-Cys Prx are present in the cytosol but differ in their expression patterns.<ref name=Usui2012>Usui M, Masuda-Suganuma H, Fukumoto S, Angeles JM, Inoue N, Kawazu SI (2012) Expression profiles of peroxiredoxins in liver stage of the rodent malaria parasite ''Plasmodium berghei''. Parasitol Int pii: S1383-5769(12)00159-6. {{DOI|10.1016/j.parint.2012.11.007}}</ref> TPx-1 is transcribed shortly after infection of the hepatocyte and expression continues until the schizont stage. Transcription of 1-Cys Prx starts after the parasite has developed into the schizont stage. |
|||
The 1-Cys peroxiredoxin enzyme appears to be located in the cytoplasm.<ref name=Hakimi2012>Hakimi H, Asada M, Angeles JM, Kawai S, Inoue N, Kawazu SI (2012) ''Plasmodium vivax'' and ''P. knowlesi'': cloning, expression and functional analysis of 1-Cys peroxiredoxin. Exp Parasitol pii: S0014-4894(12)00329-3. {{DOI|10.1016/j.exppara.2012.10.018}}</ref> The peroxiredoxin requires both glutaredoxin and glutathione for its activity.<ref name=Djuika2013>Djuika CF, Fiedler S, Schnölzer M, Sanchez C, Lanzer M, Deponte M (2013) ''Plasmodium falciparum'' antioxidant protein as a model enzyme for a special class of glutaredoxin/glutathione-dependent peroxiredoxins. Biochim Biophys Acta pii: S0304-4165(13)00148-7. {{DOI|10.1016/j.bbagen.2013.04.020}}</ref> |
|||
The crystal structure of [[thioredoxin reductase]] has been solved.<ref name=Fritz-Wolf2013>Fritz-Wolf K, Jortzik E, Stumpf M, Preuss J, Iozef R, Rahlfs S, Becker K (2013) Crystal structure of the ''Plasmodium falciparum'' thioredoxin reductase-thioredoxin complex.J Mol Biol pii: S0022-2836(13)00432-4. {{DOI|10.1016/j.jmb.2013.06.037}}</ref> There are significant differences between the human enzyme and the plasmodial which are most apparent in the ''Plasmodium''-specific insertion and the conformation of the flexible C-terminal arm. |
|||
Lysis of the erythrocyte releases [[methaemoglobin]]. Exposure of uninfected erythrocytes to methaemoglobin renders them susceptible to osmotic stress and haemolysis.<ref name=Balaji2013>Balaji SN, Trivedi V (2013) Extracellular methemoglobin mediated early ROS spike triggers osmotic fragility and RBC destruction: An insight into the enhanced hemolysis during malaria. Indian J Clin Biochem 27(2) 178-85. {{DOI|10.1007/s12291-011-0176-5}}</ref> This mechanism may form part of a chain reaction resulting in the haemolysis that is found in severe malaria. Both [[N-acetyl cysteine]] and [[mannitol]] can reduce this oxidative stress. [[Clotrimazole]] irreversibly inactivates methaemoglobin and abolishes its peroxidase activity. |
|||
[[Ferriprotoporphyrin IX]] is released inside the digestive vacuole of the malaria parasite during the digestion of host cell hemoglobin.<ref name=Ginsburg2003>Ginsburg H, Golenser J (2003) Glutathione is involved in the antimalarial action of chloroquine and its modulation affects drug sensitivity of human and murine species of ''Plasmodium''. Redox Rep 8(5) 276-279</ref> Undegraded ferriprotoporphyrin IX accumulates in the membrane fraction and is degraded by reduced glutathione in a radical mediated mechanism. |
|||
Three [[phosducin]]-like proteins have been identified in ''[[Plasmodium berghei]]''.<ref name=Putonti2012>Putonti C, Quach B, Kooistra RA, Kanzok SM (2012) The evolution and putative function of phosducin-like proteins in the malaria parasite ''Plasmodium''. Infect Genet Evol pii: S1567-1348(12)00294-8. {{DOI|10.1016/j.meegid.2012.08.023}}</ref> Their role in the parasite's metabolism has yet to be clearly established. |
|||
The type II [[NADH]]:[[ubiquinone]] [[oxidoreductase]] has been shown to be redundant in the blood forms but to be essential in the mosquito midgut.<ref name=Boysen2011>{{cite journal |author=Boysen KE, Matuschewski K |year=2011 |title=Arrested oocyst maturation in ''Plasmodium'' parasites lacking type II NADH:ubiquinone dehydrogenase |journal=J Biol Chem |doi=10.1074/jbc.M111.269399 |volume=286 |issue=37 |pages=32661–71 |pmid=21771793 |pmc=3173203}}</ref> |
|||
At least one function of B<sub>6</sub> in this parasite is as an antioxidant.<ref name="Knöckel2012">{{cite journal |author=Knöckel J, Müller IB, Butzloff S, Bergmann B, Walter RD, Wrenger C |year=2012 |title=The antioxidative effect of ''de novo'' generated vitamin B6 in ''Plasmodium falciparum'' validated by protein interference |journal=Biochem J |doi=10.1042/BJ20111542 |volume=443 |issue=2 |pages=397–405 |pmid=22242896}}</ref> |
|||
The thioredoxin like protein TRX2 is not essential but its deletion reduces growth rate.<ref name=Matthews2013>Matthews K, Kalanon M, Chisholm SA, Sturm A, Goodman CD, Dixon MW, Sanders PR, Nebl T, Fraser F, Haase S, McFadden GI, Gilson PR, Crabb BS, de Koning-Ward TF (2013) The ''Plasmodium'' translocon of exported proteins (PTEX) component thioredoxin-2 is important for maintaining normal blood-stage growth. Mol Microbiol {{DOI|10.1111/mmi.12334}}</ref> |
|||
The [[glyoxalase]] system is the main catabolic route for [[methylglyoxal]], a non-enzymatic glycolytic byproduct with toxic and mutagenic effects.<ref name=SousaSilva2012>Sousa Silva M, Ferreira AE, Gomes R, Tomás AM, Ponces Freire A, Cordeiro C (2012) The glyoxalase pathway in protozoan parasites. Int J Med Microbiol 302(4-5) 225-229 {{DOI|10.1016/j.ijmm.2012.07.005}}</ref> In ''Plasmodium falciparum'' the glyoxalase pathway is glutathione dependent. Its glyoxalase I is an atypical monomeric enzyme with two active sites. |
|||
===Calcium fluctuation=== |
|||
In recently invaded erythrocytes the Ca<sup>2+</sup> concentration increases about 10 fold.<ref name=Wasserman1990>Wasserman M, Vernot JP, Mendoza PM (1990) Role of calcium and erythrocyte cytoskeleton phosphorylation in the invasion of ''Plasmodium falciparum''. Parasitol Res 76(8) 681-688</ref> The Ca<sup>2+</sup> content increases as the parasite matures.<ref name=Tanabe1990>Tanabe K (1990) Ion metabolism in malaria-infected erythrocytes. Blood Cells 16(2-3) 437-449</ref> In infected erythrocytes, Ca<sup>2+</sup> is almost exclusively localized in the parasite compartment and changes but little in the cytosol of the host cell. |
|||
Cytosolic [[calcium]]<sup>2+</sup> increases evoked by extracellular stimuli are may be observed in the form of oscillating Ca<sup>2+</sup> spikes in eukaryotic cells. Spontaneous spikes in the calcium levels have been observed in ''Plasmodium falciparum''.<ref name=Enomoto2012>Enomoto M, Kawazu S, Kawai S, Furuyama W, Ikegami T, Watanabe J, Mikoshiba K (2012) Blockage of spontaneous Ca(2+) oscillation causes cell death in intraerythrocitic ''Plasmodium falciparum''" ''PLoS One'' 7(7) e39499.</ref> The frequency of Ca2+ oscillations are higher in early ring forms than that in early trophozoites. Blockage of this oscillation results in the cessation of intraerythrocytic maturation and death of the parasite. This effect is maximal in the trophozoites. |
|||
An [[inositol phosphate kinase]] with a role in calcium metabolism has been cloned.<ref name=Stritzke2012>Stritzke C, Nalaskowski MM, Fanick W, Lin H, Mayr GW (2012) A ''Plasmodium'' multi-domain protein possesses multiple inositol phosphate kinase activities. Mol Biochem Parasitol pii: S0166-6851(12)00249-6. {{DOI|10.1016/j.molbiopara.2012.10.005}}</ref> |
|||
Expression of the [[calmodulin]] gene is developmentally regulated throughout the blood-stage cycle.<ref name=Polson2005>Polson HE, Blackman MJ (2005) A role for poly(dA)poly(dT) tracts in directing activity of the Plasmodium falciparum calmodulin gene promoter. Mol Biochem Parasitol 141(2) 179-189</ref> |
|||
[[Phosphoinositide]]-specific [[phospholipase]] C (PI-PLC) is a major regulator of [[calcium]]-dependent signal transduction, usually by liberation of calcium from intracellular stores through the action of its product, inositol-(1,4,5)-trisphosphate. These genes are found in ''P. falciparum'' and appear to be essential.<ref name=Raabe2011>Raabe A, Berry L, Sollelis L, Cerdan R, Tawk L, Vial HJ, Billker O, Wengelnik K (2011) Genetic and transcriptional analysis of phosphoinositide-specific phospholipase C in ''Plasmodium'' Exp Parasitol</ref> The genes are twice as long as their mammalian counterparts and belong to the delta class of phospholipase C proteins. |
|||
===Nucleotide metabolism=== |
|||
''[[Plasmodium falciparum|P. falciparum]]'' is unable to biosynthesize [[purines]].<ref name="gardner"/> Instead, the parasite is able to transport and interconvert host [[purines]]. The enzyme [[hypoxanthine-guanine phosphoribosyltransferase|hypoxanthine-guanine-xanthine phosphoribosyltransferase]] converts [[hypoxanthine]] to [[inosine]] monophosphate and is essential for purine salvage.<ref name=Clinch2013>Clinch K, Crump DR, Evans GB, Hazleton KZ, Mason JM, Schramm VL, Tyler PC (2013) cyclic phosph(on)ate inhibitors of ''Plasmodium falciparum'' hypoxanthine-guanine-xanthine phosphoribosyltransferase. Bioorg Med Chem pii: S0968-0896(13)00140-5. {{DOI|10.1016/j.bmc.2013.02.016}}</ref> |
|||
Conversely, the parasite can produce [[pyrimidines]] ''de novo'' using [[glutamine]], [[bicarbonate]] and [[aspartate]].<ref name="gardner"/> |
|||
A gene encoding [[S-adenosyl-L-homocysteine hydrolase]] is present in the genome.<ref name=Tanaka2013>Tanaka N, Umeda T, Kusakabe Y, Nakanishi M, Kitade Y, T Nakamura K (2013) Structural biology for developing antimalarial compounds. Yakugaku Zasshi 133(5) 527-537</ref> |
|||
''P. falciparum'' contains both cytosolic and mitochondrial [[serine]] [[hydroxymethyltransferase]] isoforms.<ref name="Spalding2010">{{cite journal |author=Spalding MD, Allary M, Gallagher JR, Prigge ST |year=2010 |title=Validation of a modified method for Bxb1 mycobacteriophage integrase-mediated recombination in ''Plasmodium falciparum'' by localization of the H-protein of the glycine cleavage complex to the mitochondrion. Mol. Biochem |journal=Parasitol.}}</ref> This is a [[pyridoxal phosphate]] dependent enzyme which plays a vital role in the ''de novo'' [[pyrimidine]] biosynthesis pathway. Both genes are expressed throughout the erythrocytic stages.<ref name=Pornthanakasem2012>Pornthanakasem W, Kongkasuriyachai D, Uthaipibull C, Yuthavong Y, Leartsakulpanich U (2012) ''Plasmodium'' serine hydroxymethyltransferase: indispensability and display of distinct localization. Malar J 11(1) 387</ref> Both enzymes appear to be essential. |
|||
A conserved trytophan residue is involved in the activity of the [[purine nucleoside phosphorylase]] enzyme.<ref name=Suthar2013>Suthar MK, Verma A, Doharey PK, Singh SV, Saxena JK (2013) Single tryptophan of disordered loop from ''Plasmodium falciparum'' purine nucleoside phosphorylase: Involvement in catalysis and microenvironment. Appl Biochem Biotechnol</ref> |
|||
[[Orotate phosphoribosyltransferase]] catalyzes the [[magnesium]] dependent condensation of [[orotic acid]] with 5-α-D-[[phosphorylribose]] 1-diphosphate to yield diphosphate and the nucleotide [[orotidine 5'-monophosphate]]. This enzyme has been crystallised.<ref name=Takashima2012>Takashima Y, Mizohata E, Tokuoka K, Krungkrai SR, Kusakari Y, Konishi S, Satoh A, Matsumura H, Krungkrai J, Horii T, Inoue T (2012) Crystallization and preliminary X-ray diffraction analysis of orotate phosphoribosyltransferase from the human malaria parasite ''Plasmodium falciparum''. Acta Crystallogr Sect F Struct Biol Cryst Commun 68(2) 244-246</ref> |
|||
===Molecular biology=== |
|||
The long adenosine/thymidine tracts that are scattered through the genome may play a role in gene duplication.<ref name=Guler2013>Guler JL, Freeman DL, Ahyong V, Patrapuvich R, White J, Gujjar R, Phillips MA, Derisi J, Rathod PK (2013) Asexual populations of the human malaria parasite, ''Plasmodium falciparum'', use a two-step genomic strategy to acquire accurate, beneficial DNA amplifications" ''PLoS Pathog'' 9(5) e1003375. {{DOI|10.1371/journal.ppat.1003375}}</ref> |
|||
The [[centromere]]s occupy a 4-4.5 kilobase region in each chromosome.<ref name=Hoeijmakers2012>Hoeijmakers WA, Flueck C, Françoijs KJ, Smits AH, Wetzel J, Volz JC, Cowman AF, Voss T, Stunnenberg HG, Bártfai R (2012) ''Plasmodium falciparum'' centromeres display a unique epigenetic makeup and cluster prior to and during schizogony. Cell Microbiol {{doi|10.1111/j.1462-5822.2012.01803.x}}</ref> The centromeres cluster to a single nuclear location prior to and during mitosis and cytokinesis but dissociate soon after invasion. |
|||
The [[DNA polymerase]] is unusual.<ref name=Schoenfeld2013>Schoenfeld TW, Murugapiran S, Dodsworth JA, Floyd S, Lodes M, Mead DA, Hedlund BP (2013) Lateral gene transfer of Family A DNA polymerases between thermophilic viruses, Aquificae, and Apicomplexa. Mol Biol Evol</ref> They share a large amino-terminal domains with putative helicase/primase elements features that are known only in the thermophilic viruses and Aquificae. A horizont transfer seems the most likely explanation for these findings. |
|||
The [[telomerase]] (''tert'') is a large protein (2518 [[codon]]s) and has a predicted molecular weight of ~280 kilo[[Dalton (unit)|Dalton]]s.<ref name=Figueiredo2005/> It has the usual telomerase specific motifs within the N-terminal half of the protein (GQ/N, CP, QFP and T) and reverse transcriptase (RT) specific motifs in the C-terminal half. The N-terminal half is required for efficient binding of the RNA template, defining the 5′ RNA template boundary, multimerization and interactions with associated proteins. The RT domain is essential for the catalytic activity. The protein contains several nuclear localization signals and is found in the [[nucleolus]]. |
|||
A putative tyrosine site specific [[recombinase]] has been isolated.<ref name=Ghorbal2012>Ghorbal M, Scheidig-Benatar C, Bouizem S, Thomas C, Paisley G, Faltermeier C, Liu M, Scherf A, Lopez-Rubio JJ, Gopaul DN (2012) Initial characterization of the Pf-int recombinase from the malaria parasite ''Plasmodium falciparum''" ''PLoS One'' 7(10) e46507. {{DOI|10.1371/journal.pone.0046507}}</ref> The N-terminus has the typical [[alpha helix|alpha helical]] bundle and potentially a mixed alpha-beta domain resembling that of λ-Int. The C-terminal domain has the putative tyrosine recombinase conserved active site residues Lysine-Histadine-Lysine-(Histadine/Tryptophan)-Tyrosine. The gene is expressed differentially during the erythrocytic stages being maximal in the schizont stage. The open reading frame encodes a ∼57 kiloDalton protein. Knockout mutants are viable and appear normal. DNA binding studies suggest a number of targets include the subtelomeric regions. |
|||
A number of [[mini chromosome maintenance]] proteins are present in the genome.<ref name=Ansari2012>Ansari A, Tuteja R (2012) Genome wide comparative comprehensive analysis of ''Plasmodium falciparum'' MCM family with human host. Commun Integr Biol 5(6) 607-615</ref> These are large proteins and members of the AAA ATPase family with a conserved region of ~200 amino acids responsible for nucleotide binding. They are responsible for unwinding DNA at the replication forks and are involved in other chromosome transactions such as transcription, chromatin remodeling and genome stability. |
|||
A SIP2 gene, a member of the ApiAP2 family of putative transcription factors, has been cloned.<ref name=Flueck2010>Flueck C, Bartfai R, Niederwieser I, Witmer K, Alako BT, Moes S, Bozdech Z, Jenoe P, Stunnenberg HG, Voss TS (2010) A major role for the ''Plasmodium falciparum'' ApiAP2 protein PfSIP2 in chromosome end biology" ''PLoS Pathog'' 6(2) e1000784. {{DOI|10.1371/journal.ppat.1000784}}</ref> It appears to be involved in maintenance of the chromosome ends rather than in the regulation of particular genes. |
|||
A novel DNA/RNA binding protein PfAlba has been described.<ref name="Chêne2011">{{cite journal |author=Chêne A, Vembar SS, Rivière L, Lopez-Rubio JJ, Claes A, Siegel TN, Sakamoto H, Scheidig-Benatar C, Hernandez-Rivas R, Scherf A |year=2011 |title=PfAlbas constitute a new eukaryotic DNA/RNA-binding protein family in malaria parasites |journal=Nucleic Acids Res}}</ref> This protein is related to the archaeal protein Alba (Acetylation lowers binding affinity). There are at least four [[paralog]]s of the PfAlba gene and these proteins form a complex with the ''P. falciparum'' specific TARE6 (Telomere-Associated Repetitive Elements 6) subtelomeric regions. Also associated with the TARE6 regions are PfSir2 a histone deacetylase. In the early blood stages the PfAlba proteins are enriched at the nuclear periphery and associate with the PfSir2 proteins. When the parasite switches from trophozoite to the schizont stage the PfAlba proteins move to the cytoplasm. These proteins will also bind single stranded RNA but the reason for this binding is not known. |
|||
The [[Single-strand binding protein|single stranded DNA binding protein]] (SSB) plays an important role in all known organisms. A SSB protein is encoded in the genome and localises to the apicoplast.<ref name=Antony2012>Antony E, Weiland EA, Korolev S, Lohman TM (2012) ''Plasmodium falciparum'' SSB tetramer wraps single stranded DNA with similar topology but opposite polarity to ''E. coli'' SSB. J Mol Biol</ref> It forms a homo-tetramer alone and when bound to single stranded DNA. The protein binds 52-65 nucleotides/tetramer.<ref name=Antony2012a>Antony E, Kozlov AG, Nguyen B, Lohman TM (2012) ''Plasmodium falciparum'' SSB tetramer binds single stranded DNA only in a fully wrapped mode. J Mol Biol</ref> While similar in its overall structure to that of the SSB of ''[[E. coli]]'' it differs at the carboxy terminal region. Although it binds single stranded DNA in a similar fashion to the SSB of ''E. coli'' it does so with the opposite polarity. There are a number of other functional differences between this protein and that of ''E. coli''. The basis for these differences has yet to be determined. |
|||
A protein - RPA1L - is the homologue of the bacterial single stranded binding protein (SSB) and acts in initiating homologous pairing and strand exchange activity.<ref name=Gopalakrishnan2013>Gopalakrishnan AM, Kumar N (2013) Opposing roles for two molecular forms of replication protein A in Rad51-Rad54-mediated DNA recombination in ''Plasmodium falciparum''. MBio 4(3) pii: e00252-13. {{DOI|10.1128/mBio.00252-13}}</ref> It is negatively regulated in a dose dependent manner by RPA1S. |
|||
The eukaryotic homologue of the bacterial [[RecA]] protein is Rad51. The ''Plasmodium falciparum'' Rad51 protein exhibits ATPase activity and promotes DNA strand exchange.<ref name="Gopalakrishnan2013"/> This protein interacts with Rad54 and replication protein A. |
|||
SET is a conserved nuclear protein involved in chromatin dynamics.<ref name=Pace2006>Pace T, Olivieri A, Sanchez M, Albanesi V, Picci L, Siden Kiamos I, Janse CJ, Waters AP, Pizzi E, Ponzi M (2006) Set regulation in asexual and sexual ''Plasmodium'' parasites reveals a novel mechanism of stage-specific expression" ''Mol Microbiol'' 60(4) 870-882</ref> In ''P falciparum'' it is expressed in both asexual and sexual blood stages but strongly accumulates in male gametocytes. In ''P falciparum'' there are two distinct promoters upstream. One is active in all blood stages while the other is active only in gametocytes and in a fraction of schizonts possibly committed to sexual differentiation. In ookinetes both promoters exhibit a basal activity, while in the oocysts the gametocyte specific promoter is silent and the reporter gene is only transcribed from the constitutive promoter. |
|||
''Plasmodium'' appears to lack both DNA methylation and the RNA interference machinery.<ref name=Baum2009>Baum J, Papenfuss AT, Mair GR, Janse CJ, Vlachou D, ''et al''. Molecular genetics and comparative genomics reveal RNAi is not functional in malaria parasites" ''Nucleic Acids Res'' 37:3788–3798</ref><ref name=Choi2006>Choi SW, Keyes MK, Horrocks P (2006) LC/ESI-MS demonstrates the absence of 5-methyl-2′-deoxycytosine in ''Plasmodium falciparum'' genomic DNA. Mol Biochem Parasitol 150:350–352</ref> |
|||
DDX6/DOZI (development of zygote inhibited) is a member of the DEAD box family and is involved in the sexual development of the protozoan parasite.<ref name=Tarique2013>Tarique M, Ahmad M, Ansari A, Tuteja R (2013) ''Plasmodium falciparum'' DOZI, an RNA helicase interacts with eIF4E. Gene pii: S0378-1119(13)00332-6. {{DOI|10.1016/j.gene.2013.03.063}}</ref> The gene is known as PfDZ50 in ''P. falciparum''. It binds DNA and RNA and has nucleic acid dependent ATPase and RNA unwinding activities. It interacts with eIF4E mainly through domain 1 and inhibits translation. It is localized mainly in the granular bodies found throughout the cytoplasm during the asexual intraerythrocytic developmental stages. |
|||
A novel transcription factor - PREBP - has been described.<ref name=Komaki-Yasuda2013>Komaki-Yasuda K, Okuwaki M, Nagata K, Kawazu S, Kano S (2013) Identification of a novel and unique transcription factor in the intraerythrocytic stage of ''Plasmodium falciparum''" ''PLoS One'' 8(9) e74701 {{DOI|10.1371/journal.pone.0074701}}</ref> This protein has 4 KH homology domains which are found in RNA-binding or single-stranded DNA-binding proteins. PREBP is well conserved in Plasmodium species and partially conserved in phylum Apicomplexa. It acts on the pf1-cys-prx, a gene expressed in the trophozoite/schizont stages. |
|||
;DNA repair proteins |
|||
Several nucleic acid repair pathways are known. These include the [[nucleotide excision repair]], the [[mismatch repair]], the [[base excision repair]], the [[double strand break repair]] and the [[cross link repair]] pathways. DNA replication errors - base substitution mismatches and insertion-deletion loops - are primarily corrected by the mismatch repair system. |
|||
The MutL homolog (MLH) - part of the [[DNA mismatch repair]] system - has been cloned.<ref name=Tarique2011>{{cite journal |author=Tarique M, Satsangi AT, Ahmad M, Singh S, Tuteja R |year=2011 |title=''Plasmodium falciparum'' MLH is schizont stage specific endonuclease |journal=Mol Biochem Parasitol}}</ref> MLH possess [[ATPase]] and [[endonuclease]] activities. Its expression is maximal in the schizont stage. |
|||
[[Polynucleotide kinase/phosphatase]] (PNKP) is a bifunctional enzyme that can phosphorylate the 5'-OH termini and dephosphorylate the 3'-phosphate termini of [[DNA]]. It is a DNA repair enzyme involved in the processing of strand break termini, which permits subsequent repair proteins to replace missing nucleotides and rejoin broken strands. A ''P. falciparum'' gene encoding a protein with 24% homology to human PNKP has been cloned.<ref name=Siribal2011>{{cite journal |author=Siribal S, Weinfeld M, Karimi-Busheri F, Mark Glover JN, Bernstein NK, Aceytuno D, Chavalitshewinkoon-Petmitr P |year=2011 |title=Molecular characterization of ''Plasmodium falciparum'' putative polynucleotide kinase/phosphatase |journal=Mol Biochem Parasitol |doi=10.1016/j.molbiopara.2011.06.007 |volume=180 |pages=1–7 |pmid=21821066 |issue=1}}</ref> This enzyme dephosphorylates single-stranded substrates or double-stranded substrates with a short 3'-single-stranded overhang, but not double-stranded substrates that mimicked single-strand breaks. |
|||
;RNA binding proteins |
|||
The messenger RNA capping system appears to be more similar to that of fungi than to vertebrates.<ref name=Ho2001>Ho CK, Shuman S (2001) A yeast-like mRNA capping apparatus in ''Plasmodium falciparum''" ''Proc Natl Acad Sci USA'' 98(6) 3050-3035</ref> It encodes [[RNA guanylyltransferase]] (Pgt1) and [[RNA triphosphatase]] (Prt1) enzymes. The triphosphatase enzyme is a member of the fungal/viral family of metal dependent [[phosphohydrolase]]s. These are structurally and mechanistically unrelated to the cysteine phosphatase type RNA triphosphatases found in [[metazoan]]s and [[plant]]s. |
|||
A protein (PfSR1) involved in alternative [[RNA splicing|splicing]] has been described.<ref name=Eshar2012>Eshar S, Allemand E, Sebag A, Glaser F, Muchardt C, Mandel-Gutfreund Y, Karni R, Dzikowski R (2012) A novel ''Plasmodium falciparum'' SR protein is an alternative splicing factor required for the parasites' proliferation in human erythrocytes. Nucleic Acids Res</ref> It appears that regulation of this gene is essential for the parasite's normal physiology. |
|||
A number of the DExD/DExH-box containing pre-mRNA processing proteins (Prps) - Pf[[Prp2p]], Pf[[Prp5p]], Pf[[Prp16p]], Pf[[Prp22p]], Pf[[Prp28p]], Pf[[Prp43p]] and Pf[[Brr2p]] - are present in the genome.<ref name=Singh2012>Singh PK, Kanodia S, Dandin CJ, Vijayraghavan U, Malhotra P (2012) ''Plasmodium falciparum'' Prp16 homologue and its role in splicing. Biochim Biophys Acta pii: S1874-9399(12)00157-5. {{DOI|10.1016/j.bbagrm.2012.08.014}}</ref> PfPrp16p a helicase and a member of DEAH-box protein family with nine collinear sequence motifs has been cloned. It binds to RNA, hydrolyses ATP and appears to be involved in splicing. |
|||
The [[RNA polymerase]] II has an unual expansion in the C terminal domain.<ref name=Kishore2009>Kishore SP, Perkins SL, Templeton TJ, Deitsch KW (2009) An unusual recent expansion of the C-terminal domain of RNA polymerase II in primate malaria parasites features a motif otherwise found only in mammalian polymerases" ''J Mol Evol'' 68(6) 706-14. {{DOI|10.1007/s00239-009-9245-2}} PMID 19449052</ref> The primate infecting species posses a larger number of repeats that are found in either the bird or rodent species. |
|||
A cyclin (Pfcyc-1) has been cloned and its structure has been solved.<ref name=Kaushik2013>Kaushik A, Subramaniam S, Gupta D (2013) ''In silico'' characterization and molecular dynamics simulation of Pfcyc-1, a cyclin homolog of ''Plasmodium falciparum''. J Biomol Struct Dyn</ref> The protein has a typical cyclin box which consists of two tandemly repeating five-helix bundles separated by a linker hinge peptide. |
|||
The origin recognition complex subunit 1 (Orc1) associates with fSir2 in the sliencing complex.<ref name=Varunan2013>Varunan SM, Tripathi J, Bhattacharyya S, Suhane T, Bhattacharyya MK () ''Plasmodium falciparum'' origin recognition complex subunit 1 (PfOrc1) functionally complements Δsir3 mutant of Saccharomyces cerevisiae. {{DOI|10.1016/j.molbiopara.2013.08.004}}</ref> |
|||
At least seven proteins are known to be involved in the splicing process.<ref name=Hossain2013>Hossain M, Sharma S, Korde R, Kanodia S, Chugh M, Rawat K, Malhotra P (2013) Organization of ''Plasmodium falciparum'' spliceosomal core complex and role of arginine methylation in its assembly. Malar J 12(1) 333</ref> They are expressed at asexual blood stages of the parasite, show nucleo-cytoplasmic localization and form a ring like heptameric complex like other eukaryotes. The interaction of PfSMN (survival of motor neuron, Tudor domain containing protein) or PfTu-TSN (tudor domain of Tudor Staphylococcal nuclease) with the PfSmD1 protein is methylation dependent. The arginine methylase PfPRMT5 appear to interact with PfSmD1 suggesting a role for arginine methylation in the assembly of the spliceosome complex. |
|||
===Helicases=== |
|||
An unusual helicase - a homologue of Dbp5 and DDX19 from yeast and human respectively - has been cloned.<ref name=Mehta2011>Mehta J, Tuteja R (2011) A novel dual Dbp5/DDX19 homologue from ''Plasmodium falciparum'' requires Q motif for activity. Mol Biochem Parasitol 176(1) 58-63 {{DOI|10.1016/j.molbiopara.2010.12.003}}</ref> It possesses DNA and RNA unwinding, nucleic acid dependent ATPase and RNA binding activities. A Q motif is required for its activity. |
|||
Another DEAD box helicase - a homolog of Has1p from yeast which has DNA and RNA unwinding, nucleic acid dependent ATPase and RNA binding activities - has also been cloned.<ref name=Prakash2010>Prakash K, Tuteja R (2010) A novel DEAD box helicase Has1p from ''Plasmodium falciparum'': N-terminal is essential for activity. Parasitol Int 59(2) 271-277 {{DOI|10.1016/j.parint.2010.02.003}}</ref> |
|||
The [[DNA helicase]] II (''uvrD'') is a superfamily 1A helicase which plays an essential role in the mismatch repair pathway. Homologs of UvrD include the proteins PcrA and Rep. These proteins have a two domain (1 and 2) structure with each domain made of two sub-domains (1A, 1B, 2A and 2B) and a C-terminal extension. They are DNA-dependent ATPases with 3′ to 5′ helicase activity. The helicase activity is located in the N terminal domain. |
|||
The UvrD protein of ''P. falciparum'' has been cloned.<ref name=Ahmad2012>Ahmad M, Ansari A, Tarique M, Satsangi AT, Tuteja R (2012) ''Plasmodium falciparum'' UvrD helicase translocates in 3' to 5' direction, colocalizes with MLH and modulates Its activity through physical interaction" ''PLoS One'' 7(11) e49385. {{DOI|10.1371/journal.pone.0049385}}</ref> This gene (PFE0705c) is located on chromosome 5 and contains no introns. It is 4326 bases in length, encodes a protein of 1441 amino acids and has a predicted molecular weight of ~170 kiloDaltons. The two domains and their subdomains are present: The 1A domain is from amino acid 1–722; the 1B domain is from amino acid 150–464; the 2A domain is from amino acid 723–1441; and the 2B domain is from amino acid 896–1359. There is no C-terminal extension. The ATPase and helicase activity are confined to domain 1A and 1B (the N-terminal and first half of the C terminal). It is expressed in the schizont stages of intraerythrocytic development and it colocalizes with PfMLH, a protein involved in mismatch repair. Both PfDH60 - another helicase - and PfMLH are also expressed in schizont stages. |
|||
A helicase - PfH45 - of 398 amino acid residues (molecular weight 45 kiloDaltons) is a unique bipolar helicase with both the 3' to 5' and 5' to 3' directional helicase activities.<ref name=Pradhan2007>Pradhan A, Tuteja R (2007) Bipolar, dual ''Plasmodium falciparum'' helicase 45 expressed in the intraerythrocytic developmental cycle is required for parasite growth" ''J Mol Biol'' 373(2) 268-281</ref> It is expressed in all the intraerythrocytic developmental stages and has a role in translation. |
|||
A 3'-5' DNA helicase has been identified in the parasite.<ref name=Suntornthiticharoen2006>Suntornthiticharoen P, Petmitr S, Chavalitshewinkoon-Petmitr P (2006) Purification and characterization of a novel 3'-5' DNA helicase from ''Plasmodium falciparum'' and its sensitivity to anthracycline antibiotics" ''Parasitology'' 133(4) 389-398</ref> The apparent molecular weight is 90 kiloDaltons. Its activity is dependent on the presence of [[magnesium]] and ATP. |
|||
A homolog of UAP56 (U2AF65 associated protein) - a member of the DEAD box helicase family - has been cloned.<ref name=Shankar2008>Shankar J, Pradhan A, Tuteja R (2008) Isolation and characterization of ''Plasmodium falciparum'' UAP56 homolog: evidence for the coupling of RNA binding and splicing activity by site-directed mutations. Arch Biochem Biophys 478(2) 143-53 {{DOI|10.1016/j.abb.2008.07.027}}</ref> This homolog - PfU52 - contains the RNA dependent ATPase, RNA helicase and RNA binding activities. This protein is expressed in all the intraerythrocytic developmental stages of the parasite. Residues glycine 181, isoleucine 182 and arginine 206 are involved in RNA binding and this binding activity is required for its enzymatic activities. |
|||
The [[RuvB]] proteins belong to AAA+ family of enzymes which are involved in diverse cellular activities. There are at least 3 copies of this protein in the genome.<ref name=Ahmad2012>Ahmad M, Singh S, Afrin F, Tuteja R (2012) Novel RuvB nuclear ATPase is specific to intraerythrocytic mitosis during schizogony of ''Plasmodium falciparum''. Mol Biochem Parasitol</ref> The PfRuvB1 protein has considerable homology with human as well as yeast RuvB1 and contains both Walker motif A and Walker motif B.<ref name=Ahmad2013a>Ahmad M, Tuteja R (2013) ''Plasmodium falciparum'' RuvB1 is an active DNA helicase and translocates in the 5'-3' direction" ''Gene'' 515(1) 99-109 {{DOI|10.1016/j.gene.2012.11.020}}</ref> It is an ATPase and this activity increased significantly in the presence of single stranded DNA. It also has DNA helicase activity and translocates preferentially in 5' to 3' direction. It is constitutively expressed during all the stages of intraerythrocytic cycle and localizes mainly to the nucleus. |
|||
PfRuvB2 similarly has both ATPase and weak DNA helicase activities.<ref name=Ahmad2013>Ahmad M, Tuteja R (2013) |
|||
''Plasmodium falciparum'' RuvB2 translocates in 5'-3' direction, relocalizes during schizont stage and its enzymatic activities are up regulated by RuvB3 of the same complex. Biochim Biophys Acta pii: S1570-9639(13)00368-3. {{DOI|10.1016/j.bbapap.2013.10.010}}</ref> It is expressed in all the asexual intraerythrocytic developmental stages and localizes mainly in the nucleus during merozoite, ring and trophozoite stages while during schizont stage it relocalizes partially in the nucleus and partially towards cytoplasm. It interacts with PfRuvB3 and not with PfRuvB1. |
|||
RuvB3 possesses the Walker motif A, Walker motif B, sensor I and sensor II conserved motifs similar to yeast and human RuvB like proteins. It has single stranded DNA dependent ATPase activity. The protein is mainly expressed during intraerythrocytic schizont stages and localizes to the nuclear region. In the merozoite the protein relocalizes to the sub nuclear region. The PfRuvB2/PfRuvB3 complex preferentially translocates and unwinds DNA in the 5'-3' direction. |
|||
===Histones and their modifying enzymes=== |
|||
[[Histone]]s are essential for the correct packing and function of DNA in eukaryotes. Variants of these proteins are known to occur throughout eukaryotes and these are thought to play a role in epigenetic control. Although variants of H2A and H3 are common, variants of H2B and H4 are much less so. H2B.Z is an apicomplexan specific H2B variant.<ref name=Hoeijmakers2013>Hoeijmakers WA, Salcedo-Amaya AM, Smits AH, Françoijs KJ, Treeck M, Gilberger TW, Stunnenberg HG, Bártfai R (2013) H2A.Z/H2B.Z double-variant nucleosomes inhabit the AT-rich promoter regions of the ''Plasmodium falciparum'' genome. Mol Microbiol {{DOI|10.1111/mmi.12151}}</ref> H2B.Z localises to euchromatic intergenic regions throughout intraerythrocytic development and with H2A.Z may form nucleosomes. These H2A.Z and H2B.Z nucleosomes are more common in promoters over 3' intergenic regions and their occurrence is related to the promotor's strength. The presence of H2B.Z is correlated with the base composition of the underlying DNA. |
|||
Other variants of histones known to occur in this genome are H2Bv, H3.3 and CenH3.<ref name=Miao2006>Miao J, Fan Q, Cui L, Li J. The malaria parasite ''Plasmodium falciparum'' histones: organization, expression, and acetylation" ''Gene'' 369:53–65</ref> Covalent modification of the histones is well known with at least 44 different types having been described.<ref name=Trelle2009>Trelle MB, Salcedo-Amaya AM, Cohen AM, Stunnenberg HG, Jensen ON. Global histone analysis by mass spectrometry reveals a high content of acetylated lysine residues in the malaria parasite ''Plasmodium falciparum''. J Proteome Res 8:3439–3450</ref> The deacetylation and subsequent tri-methylation of lysine 9 on histone H3 (H3K9me3) as well as the recruitment of heterochromatin protein 1 are found in heterochromatic islands and are involved in the regulation of the ''var'' genes.<ref name=Flueck2009>Flueck C, Bartfai R, Volz J, Niederwieser I, Salcedo-Amaya AM, ''et al'' (2009) ''Plasmodium falciparum'' heterochromatin protein 1 marks genomic loci linked to phenotypic variation of exported virulence factors" ''PLoS Pathog'' 5:e1000569</ref> Other modifications that have been described include H3K4me3, H3K9ac, H3K14ac and H4ac.<ref name="Trelle2009"/> H2A.Z is consistently found at both 5′ and 3′ ends of the euchromatic genes. The histone acetyl transferases (Gcn5 and Esa1) that can act on H2A.Z are present in the genome.<ref name=Bischoff2010>Bischoff E, Vaquero C (2010) ''In silico'' and biological survey of transcription-associated proteins implicated in the transcriptional machinery during the erythrocytic development of ''Plasmodium falciparum''" ''BMC Genomics'' 11:34</ref> |
|||
Posttranslational covalent modifications of the histones (acetylation, methylation and phosphorylation) are known to occur. How these influence gene transcription is poorly understood. The [[14-3-3 protein]] appears to act as a phosphorlyated histone 3 reader.<ref name=Dastidar2012>Dastidar EG, Dzeyk K, Krijgsveld J, Malmquist NA, Doerig C, Scherf A, Lopez-Rubio JJ (2013) Comprehensive histone phosphorylation analysis and identification of pf14-3-3 protein as a histone h3 phosphorylation reader in malaria parasites" ''PLoS One'' 8(1) e53179. {{DOI|10.1371/journal.pone.0053179}}</ref> |
|||
A [[histone deacetylase]] (HDAC1) has been cloned.<ref name=Joshi1999>Joshi MB, Lin DT, Chiang PH, Goldman ND, Fujioka H, Aikawa M, Syin C (1999) Molecular cloning and nuclear localization of a histone deacetylase homologue in ''Plasmodium falciparum''. Mol Biochem Parasitol 99(1) 11-19</ref> The protein has 449 amino acid residues and localises to the nucleus. Its molecular weight is 50 kilo[[Dalton (unit)|Dalton]]s and it is predominantly expressed in mature asexual blood stages and in gametocytes. |
|||
Sir2A is a member of the [[sirtuin]] family of [[nicotinamide adenine dinucleotide]] dependent [[deacetylase]]s. In ''P. falciparum'' it has been has been shown to regulate the expression of surface antigens to evade the detection by host immune surveillance.<ref name=Zhu2011>{{cite journal |author=Zhu AY, Zhou Y, Khan S, Deitsch K, Hao Q, Lin H |year=2011 |title=''Plasmodium falciparum'' Sir2A preferentially hydrolyzes medium and long chain fatty acyl lysine |journal=ACS Chem Biol}}</ref> While it is a poor deacetylator of [[histone]]s it also catalyzes the hydrolysis of medium and long chain fatty acyl groups from [[lysine]] residues. Proteins are present in ''P. falciparum'' with these modifications and these can be removed by can be removed by PfSir2A ''in vitro''. This suggests that this may be its role rather than the deacetylation of histones. |
|||
During the life cycle the telomeres and telomere associated repeat elements are transcribed as long non coding RNAs.<ref name=Sierra-Miranda2012>Sierra-Miranda M, Delgadillo DM, Mancio-Silva L, Vargas M, Villegas-Sepulveda N, Martínez-Calvillo S, Scherf A, Hernández-Rivas R (2012) Two long non-coding RNAs generated from subtelomeric regions accumulate in a novel perinuclear compartment in ''Plasmodium falciparum''. Mol Biochem Parasitol</ref> They are transcribed by [[RNA polymerase]] II as single-stranded molecules. In the ring stage, these transcripts are located in a single perinuclear compartment that does not co-localize with any known nuclear subcompartment. During the schizont stage they are found at several nuclear foci. At least some of these can form stable and repetitive hairpin structure that is able to bind histones. Their function requires further elucidation. |
|||
Reversible histone modifications can cause changes in gene expression and alter the phenotype.<ref name="Sharma2013"/> |
|||
===Gene regulation=== |
|||
A number of novel DNA binding sites have been identified along the genome.<ref name=Harris2011>{{cite journal |author=Harris EY, Ponts N, Le Roch KG, Lonardi S |year=2011 |title=Chromatin-driven ''de novo'' discovery of DNA binding motifs in the human malaria parasite |journal=BMC Genomics |volume=12 |issue=1 |page=601 |doi=10.1186/1471-2164-12-601}}</ref> Their function - if any - remains to be determined. |
|||
Apetala 2 (AP2) family proteins are transcription factors that have DNA-binding domains of ~60 amino acids called AP2 domains. 27 AP2-family genes have been identified in the ''Plasmodium falciparum'' genome.<ref name=Painter2011>Painter HJ, Campbell TL, Llinas M (2011) The Apicomplexan AP2 family: integral factors regulating ''Plasmodium'' development. Mol Biochem Parasitol 176: 1–7</ref> One of these proteins appears to play a critical role in the liver stage development of the parasite.<ref name=Iwanaga2012>Iwanaga S, Kaneko I, Kato T, Yuda M (2012) Identification of an AP2-family protein that is critical for malaria liver stage development" ''PLoS One'' 7(11) e47557. {{DOI|10.1371/journal.pone.0047557}}</ref> |
|||
The transcription factor [[NF-YB]] is localised in the nucleus during the erythrocytic stages of the life cycle.<ref name=Lima2012>Lima WR, Moraes M, Alves E, Azevedo MF, Passos DO, Garcia CR (2012) The PfNF-YB transcription factor is a downstream target of melatonin and cAMP signalling in the human malaria parasite ''Plasmodium falciparum''. J Pineal Res {{DOI|10.1111/j.1600-079X.2012.01021.x}}</ref> [[Melatonin]] and cyclic adenosine monophosphate modulate the expression of NF-YB. NF-YB is also more ubiquitinated in the presence of melatonin. |
|||
The protein PfMyb1 is a transcription factor belonging to the [[tryptophan]] cluster family.<ref name=Gissot2005>{{cite journal |author=Gissot M, Briquet S, Refour P, Boschet C, Vaquero C |year=2005 |title=PfMyb1, a ''Plasmodium'' falciparum transcription factor, is required for intra-erythrocytic growth and controls key genes for cell cycle regulation |journal=J Mol Biol |volume=346 |issue=1 |pages=29–42 |doi=10.1016/j.jmb.2004.11.045 |pmid=15663925}}</ref> Inhibition of this gene reduces growth by ~40% with the mortality being concentrated at the trophozoite-schizont interface. |
|||
SAP1 has been shown to be involved in the post transcriptional control of [[liver]] stage genes.<ref name="Aly2011">{{cite journal |last1=Aly |first1=AS |last2=Lindner |first2=SE |last3=MacKellar |first3=DC |last4=Peng |first4=X |last5=Kappe |first5=SH |author-separator=, |year=2011 |title=SAP1 is a critical post-transcriptional regulator of infectivity in malaria parasite sporozoite stages |journal=Mol Microbiol |volume=79 |issue=4 |pages=929–939 |doi=10.1111/j.1365-2958.2010.07497.x |pmid=21299648}}</ref> |
|||
[[CCR4-associated factor 1]] is involved in the regulation more than 1000 genes during malaria parasite's intraerythrocytic stages.<ref name=Balu2011>{{cite journal |author=Balu B, Maher SP, Pance A, Chauhan C, Naumov AV, Andrews RM, Ellis PD, Khan SM, Lin JW, Janse CJ, Rayner JC, Adams JH |year=2011 |title=CCR4-associated factor-1 coordinates expression of ''Plasmodium falciparum'' egress and invasion proteins |journal=[[Eukaryotic Cell]] |doi=10.1128/EC.05099-11 |volume=10 |issue=9 |pages=1257}}</ref> Mutations in this gene result in mistimed expression, aberrant accumulation and localization of proteins involved in parasite egress and invasion of new host cells. This leads to the premature release of predominantly half-finished merozoites in turn drastically reducing the intraerythrocytic growth rate of the parasite. |
|||
===Mutation rates=== |
|||
The overall mutation rate of the genome is 1.0-9.7 x 10<sup>-9</sup> mutations per base pair per generation.<ref name=Bopp2013>Bopp SE, Manary MJ, Bright AT, Johnston GL, Dharia NV, Luna FL, McCormack S, Plouffe D, McNamara CW, Walker JR, Fidock DA, Denchi EL, Winzeler EA (2013) Mitotic evolution of ''Plasmodium falciparum'' shows a stable core genome but recombination in antigen families" ''PLoS Genet'' 9(2) e1003293. {{DOI|10.1371/journal.pgen.1003293}}</ref> The rate in genes involved in immune evasion is higher - 9.5 x 10<sup>-6</sup> structural variants per base pair per generation. |
|||
===Protein metabolism=== |
|||
It has been hypothesized that the parasite obtains all, or nearly all, of its [[amino acids]] by salvaging from the host or through the degradation of [[hemoglobin]]. This is supported by the fact that genomic analysis has found no enzymes necessary for [[amino acid]] biosynthesis, except for glycine-serine, cysteine-alanine, aspartate-asparagine, proline-ornithine, and [[Glutamate-glutamine cycle|glutamine-glutamate interconversions]].<ref name="gardner"/> |
|||
[[Deoxyhypusine monooxygenase|Deoxyhypusine hydroxylase]] catalyses the final stage of the synthesis of the amino acid [[hypusine]]. In eukaryotes this amino acid is only found in [[eukaryotic initiation factor]] 5A. This gene has been identified in the genome of ''P. falciparum'', ''P. knowlesi'', ''P. vivax'' and ''P. yoelii''.<ref name=Atemnkeng2013>Atemnkeng VA, Pink M, Schmitz-Spanke S, Wu XJ, Dong LL, Zhao KH, May C, Laufer S, Langer B, Kaiser A (2013) Deoxyhypusine hydroxylase from ''Plasmodium vivax'', the neglected human malaria parasite: molecular cloning, expression and specific inhibition by the 5-LOX inhibitor zileuton" ''PLoS One'' 8(3) e58318. {{DOI|10.1371/journal.pone.0058318}}</ref> |
|||
[[Deoxyhypusine synthase]] catalyzes the first step in [[hypusine]] biosynthesis of [[eukaryotic initiation factor]] 5A ([[EIF5A]]). Inhibitors of this enzyme may be of use therapeutically.<ref name=Schwentke2012>Schwentke A, Krepstakies M, Müller AK, Hammerschmidt C, Motaal B, Bernhard T, Hauber J, Kaiser A (2012) ''In vitro'' and ''in vivo'' silencing of plasmodial dhs and eIF-5A genes in a putative, non-canonical RNAi-related pathway. BMC Microbiol 12(1) 107</ref> |
|||
Much of the digestion of haemoglobin occurs within the digestive vacuole. Multiple enzymes are involved in this process including four distinct plasmepsins (aspartic acid proteases). |
|||
===Protein translation=== |
|||
There are two [[EF-G|translation elongation factor G]] proteins encoded in the genome.<ref name=Johnson2011>{{cite journal |author=Johnson RA, McFadden GI, Goodman CD |year=2011 |title=Characterization of two malaria parasite organelle translation elongation factor g proteins: the likely targets of the anti-malarial fusidic acid |pmid=21695207 |doi=10.1371/journal.pone.0020633 |journal=PLoS ONE |volume=6 |issue=6 |page=e20633 |pmc=3112199 |editor1-last=Langsley |editor1-first=Gordon}}</ref> One is located in the mitochondrion and the second in the plastid. Both appear to be inhibitable with [[fusidic acid]] |
|||
Two [[Ribosome Recycling Factor]]s (RRF1 and RR2) are present in the genome.<ref name=Gupta2013>Gupta A, Mir SS, Jackson KE, Lim EE, Shah P, Sinha A, Siddiqi MI, Ralph SA, Habib S (2013) Recycling factors for ribosome disassembly in the apicoplast and mitochondrion of ''Plasmodium falciparum''. Mol Microbiol {{DOI|10.1111/mmi.12230}}</ref> Both proteins are targeted to both the apicoplast and the mitochondrion. RRF2 is also present in the cytoplasm. Unusually it forms dimers. RRF1 has a 108 amino acid insert compared with that of other organisms. The function of this insert - if any - is currently unknown. |
|||
The [[eIF2|eukaryotic translation initiation factor]] 2α has a regulatory [[serine]] at position 51. This can be phosphorylated by several kinases. Three are known in ''P falciparum'': IK1, IK2 and PK4.<ref name=Zhang2012>Zhang M, Mishra S, Sakthivel R, Rojas M, Ranjan R, Sullivan WJ Jr, Fontoura BM, Ménard R, Dever TE, Nussenzweig V (2012) PK4, a eukaryotic initiation factor 2α(eIF2α) kinase, is essential for the development of the erythrocytic cycle of ''Plasmodium''. Proc Natl Acad Sci USA</ref> IK1 regulates stress response to amino acid starvation; IK2 inhibits development of malaria sporozoites present in the mosquito salivary glands; and PK4 is essential for the completion of the parasite's erythrocytic cycle. |
|||
Unlike other organisms, ''Plasmodium'' codon bias is not correlated to tRNA gene copy number.<ref name=Filisetti2013>Filisetti D, Theobald-Dietrich A, Mahmoudi N, Rudinger-Thirion J, Candolfi E, Frugier M (2013) Aminoacylation of ''Plasmodium falciparum'' tRNAAsn and insights in the synthesis of asparagine repeats. J Biol Chem 2013</ref> |
|||
[[Aminoacyl tRNA synthetase]]s are required for protein synthesis. [[Alanine—tRNA ligase|Alanine tRNA synthetase]], [[Glycine—tRNA ligase|glycine tRNA synthetase]] and [[Threonine—tRNA ligase|threonine tRNA synthetase]] are dually localised to the cytosol and the apicoplast.<ref name=Jackson2011>{{cite journal |author=Jackson KE, Pham JS, Kwek M, De Silva NS, Allen SM, Goodman CD, McFadden GI, de Pouplana LR, Ralph SA |year=2011 |title=Dual targeting of aminoacyl-tRNA synthetases to the apicoplast and cytosol in ''Plasmodium falciparum'' |journal=Int J Parasitol}}</ref> These enzymes do not appear to be present in the mitochondrion. |
|||
[[YARS|Tyrosyl tRNA synthetase]] is secreted by the parasite into the [[cytoplasm]] of the infected erythrocyte.<ref name=Bhatt2011>Bhatt TK, Khan S, Dwivedi VP, Banday MM, Sharma A, Chandele A, Camacho N, de Pouplana LR, Wu Y, Craig AG, Mikkonen AT, Maier AG, Yogavel M, Sharma A (2011) Malaria parasite tyrosyl-tRNA synthetase secretion triggers pro-inflammatory responses. Nat Commun 2:530. {{doi|10.1038/ncomms1522}}</ref> On lysis of the erythrocyte it is released into the blood stream where it is pro inflammatory. It is specifically bound by and taken up by host macrophages and leads to enhanced secretion of the [[cytokine]]s [[tumor necrosis factor-alpha]] and [[interleukin 6]]. This interaction also increases the adherence linked host endothelial receptors [[ICAM-1]] and [[VCAM-1]]. |
|||
The cytoplasmic [[KARS (gene)|lysyl-tRNA synthetase]] is dimeric unlike the human version which may be dimeric or tetrameric.<ref name=Khan2013>Khan S, Garg A, Camacho N, Van Rooyen J, Kumar Pole A, Belrhali H, Ribas de Pouplana L, Sharma V, Sharma A (2013) Structural analysis of malaria-parasite lysyl-tRNA synthetase provides a platform for drug development. Acta Crystallogr D Biol Crystallogr 69(5) 785-795 {{DOI|10.1107/S0907444913001923}}</ref> It is capable of synthesizing the signalling molecule [[Ap4A|diadenosine tetraphosphate]] using ATP as a substrate. |
|||
The [[WARS (gene)|tryptophanyl-tRNA synthetase]] appears to be localised to the cytoplasm.<ref name=Khan2013>Khan S, Garg A, Sharma A, Camacho N, Picchioni D, Saint-Léger A, Ribas de Pouplana L, Yogavel M, Sharma A (2013) An appended domain results in an unusual architecture for malaria parasite tryptophanyl-tRNA synthetase" ''PLoS One'' 8(6) e66224. {{DOI|10.1371/journal.pone.0066224}}</ref> This enzyme has an unusual N-terminal extension that appears to be essential for its activity. |
|||
===Post translational modifications=== |
|||
Palmitoylation - the reversible addition of a [[lipid]] moiety to a [[cysteine]] residue - appears to be common in this parasite.<ref name=Jones2012>Jones ML, Collins MO, Goulding D, Choudhary JS, Rayner JC (2012) Analysis of protein palmitoylation reveals a pervasive role in ''Plasmodium'' development and pathogenesis. Cell Host Microbe 12(2) 246-258</ref> Its role in its biology is not yet understood. |
|||
An N-[[myristoyltransferase]] is present in the genome.<ref name=Rackham2012>Rackham MD, Brannigan JA, Moss DK, Yu Z, Wilkinson AJ, Holder AA, Tate EW, Leatherbarrow RJ (2012) Discovery of novel and ligand-efficient inhibitors of ''Plasmodium falciparum'' and ''Plasmodium vivax'' N-Myristoyltransferase. J Med Chem</ref> It is involved in protein trafficking. |
|||
Lysine acetylation appears to be common in this organism.<ref name=Miao2013>Miao J, Lawrence M, Jeffers V, Zhao F, Parker D, Ge Y, Sullivan WJ Jr, Cui L (2013) Extensive lysine acetylation occurs in evolutionarily conserved metabolic pathways and parasite-specific functions during ''Plasmodium falciparum'' intraerythrocytic development. Mol Microbiol {{DOI|10.1111/mmi.12303}}</ref> Lysine-acetylated proteins are present in the nucleus, cytoplasm, mitochondrion and apicoplast. The acetyltransferase PfMYST appears to be involved in this process. The effects of these modifications are not yet understood. |
|||
==Metabolism== |
|||
While all of the metabolic pathways of [[Plasmodium falciparum]] have yet to be fully elucidated, the presence and components of many can be predicted through genomic analysis.<ref name="gardner"/> |
|||
===Hemoglobin metabolism=== |
===Hemoglobin metabolism=== |
||
During the erythrocytic stage of the parasite's life cycle, it uses intracellular [[hemoglobin]] as a food source. The protein is broken down into peptides, and the heme group is released and detoxified by [[biocrystallization]] in the form of hemozoin. |
|||
During the erythrocytic stage of the parasite's life cycle, it uses intracellular [[hemoglobin]] as a food source. The protein is broken down into peptides, and the heme group is released and detoxified by [[biocrystallization]] in the form of hemozoin.<ref>{{cite journal |author=Hempelmann E. |title=Hemozoin biocrystallization in Plasmodium falciparum and the antimalarial activity of crystallization inhibitors |journal=Parasitol Research |volume=100 |issue=4 |pages=671–676 |year=2007 |pmid=17111179 |doi=10.1007/s00436-006-0313-x |url=http://parasitology.informatik.uni-wuerzburg.de/login/n/h/j_436-100-4-2006-11-17-313.html}}{{dead link|date=October 2013}}</ref> |
|||
[[Heme]] biosynthesis by the parasite has been reported.<ref name="bonday">{{cite journal |
[[Heme]] biosynthesis by the parasite has been reported.<ref name="bonday">{{cite journal |
||
|last=Bonday |
|||
|first=Z.Q. |
|||
|title=Import of host delta-aminolevulinate dehydratase into the malarial parasite: Identification of a new drug target |
|||
| authorlink = |
|||
|journal=Nature Medicine |
|||
| coauthors = |
|||
|volume=6 |
|||
| title = Import of host delta-aminolevulinate dehydratase into the malarial parasite: Identification of a new drug target |
|||
|issue=8 |
|||
| journal = Nature Medicine |
|||
|pages=898–903 |
|||
| volume = 6 |
|||
|year=2002 |
|||
| issue = |
|||
|url=http://www.nature.com/nm/journal/v6/n8/full/nm0800_898.html#B1 |
|||
| pages = 898–903 |
|||
|doi=10.1038/78659 |
|||
| publisher = |
|||
|pmid=10932227 |
|||
| location = |
|||
|last2=Dhanasekaran |
|||
| date = 2002 |
|||
|first2=S |
|||
| url = http://www.nature.com/nm/journal/v6/n8/full/nm0800_898.html#B1 |
|||
|last3=Rangarajan |
|||
| doi =10.1038/78659 |
|||
|first3=PN |
|||
| id = |
|||
|last4=Padmanaban |
|||
| accessdate = }}</ref> |
|||
|first4=G}}</ref> Mutations have been induced in both the first and last enzymes (δ-[[aminolevulinate synthase]] and [[ferrochelatase]]) of this pathway.<ref name=Nagaraj2013>Nagaraj VA, Sundaram B, Varadarajan NM, Subramani PA, Kalappa DM, Ghosh SK, Padmanaban G (2013) Malaria parasite-synthesized heme is essential in the mosquito and liver stages and complements host heme in the blood stages of infection. PloS Pathog (8) e1003522. {{DOI|10.1371/journal.ppat.1003522}}</ref> Although these mutations do not appear to have an effect on the growth in blood cells, growth in both the mosquito and in the liver requires an intact pathway. The muatated strains form decreased numbers of oocyts and fail to form sporozoites. These strains fail to be infective to hosts. The parasites incorporate both host haemoglobin heme and parasite synthesized heme into haemozoin and into the mitochondrial [[cytochrome]]s. |
|||
===Haemozoin=== |
|||
[[Hemozoin|Haemozoin]] (malarial pigment) is a [[ferriprotoporphyrin]] IX crystal produced by ''Plasmodium'' parasites after haemoglobin catabolism. It is an insoluble phase of iron (III) [[protoporphyrin]]-IX. The structure of haemozoin was solved in 2000 by powder X-ray diffraction.<ref name=Bohle2012>Bohle DS, Dodd EL, Stephens PW (2012) Structure of malaria pigment and related propanoate-linked metalloporphyrin dimers. Chem Biodivers 9(9) 1891-1902 {{DOI|10.1002/cbdv.201200033}}</ref> It is a crystalline structure composed of [[heme]] units interlinked to form cyclic dimers via reciprocal [[iron]]-[[oxygen]] (propionate) bonds.<ref name=Kapishnikov2012>Kapishnikov S, Berthing T, Hviid L, Dierolf M, Menzel A, Pfeiffer F, Als-Nielsen J, Leiserowitz L (2012) Aligned hemozoin crystals in curved clusters in malarial red blood cells revealed by nanoprobe X-ray Fe fluorescence and diffraction. Proc Natl Acad Sci USA</ref> Its nucleation occurs at the inner membrane of the digestive vacuole, with crystallization occurring in the aqueous rather than lipid phase.<ref name="Kapishnikov2012"/> Acylglycerol lipids are involved in the nucleation process. |
|||
[[Ferriprotoporphyrin IX]] (haematin) competes with [[NADH]] for the active site of the enzyme [[lactate dehydrogenase]].<ref name=Cortopassi2011>Cortopassi WA, Oliveira AA, Guimaraes AP, Renno MN, Krettli AU, Franca TC (2011) Docking studies on the binding of quinoline derivatives and hematin to ''Plasmodium falciparum'' lactate dehydrogenase. J Biomol Struct Dyn 29(1) 207-218</ref> This competition may be fatal to the parasite. To detoxify the ferriprotoporphyrin IX the parasite polymerizes haematin to haemozoin. |
|||
The conversion of haemoglobin to haemozoin is mediated by a ~200 kiloDalton protein complex within the digestive vacuole.<ref name=Chugh2013>Chugh M, Sundararaman V, Kumar S, Reddy VS, Siddiqui WA, Stuart KD, Malhotra P (2013) Protein complex directs hemoglobin-to-hemozoin formation in ''Plasmodium falciparum''. Proc Natl Acad Sci USA</ref> This complex contains a number of proteins including falcipain 2/2', plasmepsin II, plasmepsin IV, histo-aspartic protease and heme detoxification protein. The proteins spontaneously associate with each other and can convert haemoglobin to haemozoin. The heme detoxification protein has two heme-binding sites.<ref name=Nakatani2013>Nakatani K, Ishikawa H, Aono S, Mizutani Y (2013) Heme-binding properties of heme detoxification protein from ''Plasmodium falciparum''. Biochem Biophys Res Commun pii: S0006-291X(13)01458-7. {{DOI|10.1016/j.bbrc.2013.08.100}}</ref> The drugs chloroquine and artemisinin both act during the heme polymerization step. Chloroquine also acts at the haemoglobin degradation step. |
|||
Although the parasite has a heme oxygenase like protein which binds bound heme and protoporphyrin IX with modest affinity, this protein does not catalyze heme degradation ''in vivo''.<ref name=Sigala2012>Sigala PA, Crowley JR, Hsieh S, Henderson JP, Goldberg DE (2012) Direct tests of enzymatic heme degradation by the malaria parasite ''Plasmodium falciparum''. J Biol Chem</ref> It lacks a critical heme-coordinating [[histadine]] residue and this probably accounts for its lack of heme oxygenase activity. The function of this protein remains unknown. |
|||
;Drugs interfering with haemozoin formation |
|||
[[Artemisinin]] an anti malarial agent appears to require haemoglobin digestion for its activity.<ref name=Klonis2011>{{cite journal |author=Klonis N, Crespo-Ortiz MP, Bottova I, Abu-Bakar N, Kenny S, Rosenthal PJ, Tilley L |year=2011 |title=Artemisinin activity against ''Plasmodium falciparum'' requires hemoglobin uptake and digestion |journal=Proc Natl Acad Sci U S A |doi=10.1073/pnas.1104063108 |volume=108 |issue=28 |pages=11405–10 |pmid=21709259 |pmc=3136263}}</ref> Inhibition of hemoglobinase activity with cysteine protease inhibitors, knockout of [[falcipain]] 2 or deprivation of host cell lysate reduce the activity of the drug against the parasite. |
|||
[[Chloroquine]] causes a dose dependent decrease in haemozoin by disrupting hemozoin crystal growth.<ref name=Combrinck2012>Combrinck JM, Mabotha TE, Ncokazi KK, Ambele MA, Taylor D, Smith PJ, Hoppe HC, Egan TJ (2012) Insights into the role of heme in the mechanism of action of ntimalarials. ACS Chem Biol</ref> This results in mosaic boundaries in the crystals formed in the parasite. This disruption causes a redistribution of heme from the parasite digestive vacuole to the its cytoplasm. |
|||
Other drugs that inhibit haemozoin formation include [[amodiaquine]], [[artesunate]], [[lumefantrine]], [[mefloquine]] and [[quinine]].<ref name="Combrinck2012"/> Neither [[pyrimethamine]] nor [[sulfadoxine]] affect haemazoin formation. |
|||
;Effects on the host |
|||
Haemozoin has multiple effects on the host's immune system including increasing the level of [[matrix metalloproteinase]]-9.<ref name=Prato2011>Prato M, D'Alessandro S, Van den Steen PE, Opdenakker G, Arese P, Taramelli D, Basilico N (2011) Natural haemozoin modulates matrix metalloproteinases and induces morphological changes in human microvascular endothelium. Cell Microbiol. {{doi|10.1111/j.1462-5822.2011.01620.x}}</ref> Its presence within the lung is correlated with lung inflammation and it may be important in the pathogensis of [[acute respiratory distress syndrome]] in malaria.<ref name=Deroost2013>Deroost K, Tyberghein A, Lays N, Noppen S, Schwarzer E, Vanstreels E, Komuta M, Prato M, Lin JW, Pamplona A, Janse CJ, Arese P, Roskams T, Daelemans D, Opdenakker G, Van den Steen PE (2013) Hemozoin induces lung inflammation and correlates with malaria-associated acute respiratory distress syndrome. Am J Respir Cell Mol Biol</ref> |
|||
Haemozoin has been shown to promote inflammation mediated lysozyme release from human monocytes through p38 mitogen activated protein kinase and nuclear factor κB dependent mechanisms.<ref name=Polimeni2013>Polimeni M, Valente E, Aldieri E, Khadjavi A, Giribaldi G, Prato M (2013) Role of 15-hydroxyeicosatetraenoic acid in hemozoin-induced lysozyme release from human adherent monocytes. Biofactors {{DOI|10.1002/biof.1071}}</ref> This process may also involve [[15-hydroxyeicosatetraenoic acid]]. |
|||
Both [[matrix metalloproteinase]] 9 and its endogenous inhibitor - [[tissue inhibitor of metalloproteinase 1]] - are induced in macrophages by haemozoin.<ref name=Polimeni2013>Polimeni M, Valente E, Ulliers D, Opdenakker G, Van den Steen PE, Giribaldi G, Prato M (2013) Natural haemozoin induces expression and release of human monocyte tissue inhibitor of metalloproteinase-1" ''PLoS One'' 8(8) e71468. {{DOI|10.1371/journal.pone.0071468}}</ref> Both p38 MAPK and NF-κB appear to be involved in this induction pathway. |
|||
Haemazoin is recognised by a number of immune receptors including the [[Toll-like receptor]]s.<ref name=Tyberghein2013>Tyberghein A, Deroost K, Schwarzer E, Arese P, Van den Steen PE (2013) Immunopathological effects of malaria pigment or hemozoin and other crystals. Biofactors {{DOI|10.1002/biof.1119}}</ref> Phagolysosomal formation within the macrophages after haemozoin ingestion is normal but the haemozoin remains stored inside these cells for months or even longer without any detectable degradation. |
|||
Haemozoin inhibts erythropoetin induced proliferation of erythroid precursors.<ref name=Thawani2013>Thawani N, Tam M, Bellemare MJ, Bohle DS, Olivier M, de Souza JB, Stevenson MM (2013) Haemozoin inhibts erythropoetin induced proliferation of erythroid precursors. J Infect Dis</ref> The mechanism is unknown but this probably contributes to the anemia associated with malaria. |
|||
===Carbohydrate metabolism=== |
===Carbohydrate metabolism=== |
||
During erythrocytic stages, the parasite produces its energy mainly through anaerobic glycolysis, with [[pyruvate]] being converted into [[lactic acid|lactate]].<ref name="gardner"/> |
During erythrocytic stages, the parasite produces its energy mainly through anaerobic glycolysis, with [[pyruvate]] being converted into [[lactic acid|lactate]].<ref name="gardner"/> |
||
Genes encoding for the [[TCA cycle]] enzymes are present in the genome, but it is unclear whether the [[TCA cycle]] is used for oxidation of glycolytic products to be used for energy production, or for metabolite intermediate biosynthesis.<ref name="gardner"/> |
Genes encoding for the [[TCA cycle]] enzymes are present in the genome, but it is unclear whether the [[TCA cycle]] is used for oxidation of glycolytic products to be used for energy production, or for metabolite intermediate biosynthesis.<ref name="gardner"/> It has been hypothesized that the main function of the [[TCA cycle]] in ''[[Plasmodium falciparum|P. falciparum]]'' is for production of succinyl-CoA, to be used in heme biosynthesis.<ref name="gardner"/> |
||
Genes for nearly all of the [[pentose phosphate pathway]] enzymes have been identified from the genome sequence. |
Genes for nearly all of the [[pentose phosphate pathway]] enzymes have been identified from the genome sequence. |
||
[[Glycerol]] is a major glucose metabolite.<ref name=Lian2009>Lian LY, Al-Helal M, Roslaini AM, Fisher N, Bray PG, Ward SA, Biagini GA (2009) Glycerol: an unexpected major metabolite of energy metabolism by the human malaria parasite. Malar J 8:38. {{DOI|10.1186/1475-2875-8-38}}</ref> |
|||
===Protein metabolism=== |
|||
It has been hypothesized that the parasite obtains all, or nearly all, of its [[amino acids]] by salvaging from the host or through the degradation of [[hemoglobin]]. This is supported by the fact that genomic analysis has found no enzymes necessary for [[amino acid]] biosynthesis, except for glycine-serine, cysteine-alanine, aspartate-asparagine, proline-ornithine, and [[Glutamate-glutamine cycle|glutamine-glutamate interconversions]].<ref name="gardner"/> |
|||
Pools of several sugars (UDP-glucose, UDP-galactose, UDP-N-acetylglucosamine, GDP-mannose and GDP-fucose) are present in the cytoplasm of the blood stages.<ref name=Sanz2013>Sanz S, Bandini G, Ospina D, Bernabeu M, Mariño K, Fernández-Becerra C, Izquierdo L (2013) Biosynthesis of GDP-fucose and other sugar nucleotides in the blood-stages of ''Plasmodium falciparum''. J Biol Chem</ref> The enzymes [[GDP-mannose 4,6-dehydratase]] and [[GDP-L-fucose synthase]] are present in the genome. |
|||
Both the asexual and gametocyte stages of the life cycle utilise the [[Kreb's cycle]] to generate [[adenosine triphosphate]].<ref name=Macrae2013>Macrae JI, Dixon MW, Dearnley MK, Chua HH, Chambers JM, Kenny S, Bottova I, Tilley L, McConville MJ (2013) Mitochondrial metabolism of sexual and asexual blood stages of the malaria parasite ''Plasmodium falciparum''. BMC Biol 11(1) 67</ref> [[Glutamine]] is the preferred substrate over [[glucose]] in the asexual stages while in the gametocytes, glucose is preferred. Inhibition of this cycle in the asexual stages has little effect while inhibition in the gametocytes is lethal. |
|||
The enzyme [[hexokinase]] limits the rate of entry of glucose into [[glycolysis]].<ref name=Tjhin2013>Tjhin ET, Staines HM, van Schalkwyk DA, Krishna S, Saliba KJ (2013) Studies with the ''Plasmodium falciparum'' hexokinase reveal that PfHT limits the rate of glucose entry into glycolysis. FEBS Lett pii: S0014-5793(13)00618-2. {{DOI|10.1016/j.febslet.2013.07.052}}</ref> |
|||
Mutations in the [[glycerol kinase]] gene limit the production of [[glycerol-3-phosphate]] which is used in phosphlipid biosynthesis.<ref name=Naidoo2013>Naidoo K, Coetzer TL (2013) Reduced intra-erythrocytic growth of glycerol kinase knockout ''Plasmodium falciparum'' parasites during ''in vitro'' blood stage development. Biochim Biophys Acta pii: S0304-4165(13)00350-4. {{DOI|10.1016/j.bbagen.2013.08.006}}</ref> Exogenous [[glycerol]] may be used as an alternative source. |
|||
===Lipid metabolism=== |
===Lipid metabolism=== |
||
===Nucleotide metabolism=== |
|||
''[[Plasmodium falciparum|P. falciparum]]'' is unable to biosynthesize [[purines]].<ref name="gardner"/> Instead, the parasite is able to transport and interconvert host [[purines]]. |
|||
[[Phosphatidylcholine]] is a major and essential membrane phospholipid in the parasite. Its synthesis occurs via the CDP-choline and the serine decarboxylase phosphoethanolamine methylation pathways. The substrates of these pathways are the host's [[choline]], [[serine]] and fatty acids. Both pathways share the final two steps catalyzed by two essential enzymes [[CTP:phosphocholine cytidylyltransferase]] and [[choline-phosphate transferase]]. Mutations in [[phosphoethanolamine N-methyltransferase]] are associated with severe alterations in gametocyte development, are incapable of producing mature-stage gametocytes and are not transmittable to mosquitoes.<ref name=Bobenchik2013>Bobenchik AM, Witola WH, Augagneur Y, Nic Lochlainn L, Garg A, Pachikara N, Choi JY, Zhao YO, Usmani-Brown S, Lee A, Adjalley SH, Samanta S, Fidock DA, Voelker DR, Fikrig E, Ben Mamoun C (2013) ''Plasmodium falciparum'' phosphoethanolamine methyltransferase is essential for malaria transmission. Proc Natl Acad Sci USA </ref> |
|||
Conversely, the parasite can produce [[pyrimidines]] ''de novo'' using glutamine, bicarbonate, and aspartate.<ref name="gardner"/> |
|||
The major source of phosphatidylcholine is the CDP-choline Kennedy pathway.<ref name=Sen2013>Sen P, Vial HJ, Radulescu O (2013) Kinetic modelling of phospholipid synthesis in ''Plasmodium knowlesi'' unravels crucial steps and relative importance of multiple pathways. BMC Syst Biol 7(1):123</ref> Both phosphoethanolamine-Nmethyltransferase and phosphatidylethanolamine-N-methyltransferase are important in its synthesis. Most of the serine derived phosphatidylethanolamine is formed via serine decarboxylation, whereas the majority of phosphatidylserine is formed by base exchange reactions. |
|||
The other major component of the parasite's membranes is the phospholipid [[phosphatidylethanolamine]].<ref name=Maheshwari2012>Maheshwari S, Lavigne M, Contet A, Alberge B, Pihan E, Kocken C, Wengelnik K, Douguet D, Vial H, Cerdan R (2012) Biochemical characterization of ''Plasmodium falciparum'' CTP:phosphoethanolamine cytidylyltransferase shows that only one of the two cytidylyltransferase domains is active. Biochem J</ref> This phophlipid is not found in the erythrocyte. Phosphatidylethanolamine is synthesised ''de novo'' by the CDP-ethanolamine dependent Kennedy pathway. The rate limiting step in this pathway is the step involving CTP:phosphoethanolamine cytidylyltransferase. This enzyme is composed of two [[cytidylyltransferase]] domains separated by a linker region. Only the N-terminal domain appears to be enzymatically active. The enzyme appears to function as a dimer and to obey Michaelis-Menten kinetics. |
|||
A [[phosphatidylserine decarboxylase]] has been cloned from the parasite ''[[Plasmodium knowlesi]]''.<ref name=Choi2011>{{cite journal |author=Choi JY, Augagneur Y, Ben Mamoun C, Voelker DR |year=2011 |title=Identification of a gene encoding ''Plasmodium knowlesi'' phosphatidylserine decarboxylase by genetic complementation in yeast, and characterization of ''in vitro'' maturation of the encoded enzyme |journal=J Biol Chem}}</ref> It seems highly probably this enzyme is also found in ''P. falciparum''. |
|||
[[β-hydroxyacyl-acyl carrier protein dehydratase]] catalyzes the third and important reaction of the fatty acid elongation cycle. The ''P. falciparum'' gene has been cloned and the crystal structure of the enzyme solved.<ref name=Maity2011>Maity K, Venkata BS, Kapoor N, Surolia N, Surolia A, Suguna K (2011) Structural basis for the functional and inhibitory mechanisms of β-hydroxyacyl-acyl carrier protein dehydratase (''FabZ'') of ''Plasmodium falciparum''. J Struct Biol</ref> |
|||
α-[[lipoic acid]] (6,8-thioctic acid: LA) is a vital co-factor of α-[[ketoacid dehydrogenase]] complexes and the [[glycine]] cleavage system.<ref name=Storm2012>Storm J, Müller S (2012) lipoic acid metabolism of ''Plasmodium'' - A suitable drug target. Curr Pharm Des</ref> It is essential for the erythrocytic and liver stages of ''Plasmodium'' and is the co-factor of the acetyltransferase subunit of [[pyruvate dehydrogenase]] located in the apicoplast. |
|||
LA biosynthesis, comprising [[octanoyl-acyl carrier protein]] (ACP): protein N-[[octanoyltransferase]] and [[lipoate synthase]] is exclusively found in the apicoplast where it generates LA ''de novo'' from octanoyl-ACP, provided by the type II fatty acid biosynthesis pathway which is also present in this organelle.<ref name="Storm2012"/> |
|||
Other members of the fatty acid synthesis type II pathway present in the genome are PfFabI, PfFabG and PfFabZ.<ref name=Lauinger2013>Lauinger IL, Vivas L, Perozzo R, Stairiker C, Tarun A, Zloh M, Zhang X, Xu H, Tonge PJ, Franzblau SG, Pham DH, Esguerra CV, Crawford AD, Maes L, Tasdemir D (2013) Potential of lichen secondary metabolites against ''Plasmodium'' liver stage parasites with FAS-II as the potential target. J Nat Prod 76(6) 1064-1070</ref> |
|||
Two [[lipoic acid protein ligase]]s (LplA1 and LplA2) are present in the genome.<ref name="Storm2012"/> LplA1 is confined to the mitochondrion while LplA2 is found in both the mitochondrion and the apicoplast. LplA1 exclusively uses salvaged LA and lipoylates [[α-ketoglutarate dehydrogenase]], [[branched chain α-keto acid dehydrogenase]] and the H-protein of the glycine cleavage system. LplA2 cannot compensate for the loss of LplA1 function during blood stage development. |
|||
The parasite syntheses [[2-C-methyl-d-erythritol-4-phosphate]]. Other enzymes in the this pathway include 1-deoxy-D-xylulose 5-phosphate reductoisomerase, [[4-diphosphocytidyl-2-C-methyl-D-erythritol synthase|2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase]] and 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate synthase and [[DXP synthase|1-deoxy-D-xylulose 5-phosphate synthase]] (the first enzyme in the pathway).<ref name=Singh2013>Singh VK, Ghosh I (2013) Methylerythritol phosphate pathway to isoprenoids: Kinetic modeling and in silico enzyme inhibitions in ''Plasmodium falciparum''. FEBS Lett pii: S0014-5793(13)00482-1. {{DOI|10.1016/j.febslet.2013.06.024}}</ref> This pathway is responsible for the production of the essential isoprenoid precursors, [[isopentenyl diphosphate]] and [[dimethylallyl diphosphate]]. |
|||
The enzyme [[1-deoxy-D-xylulose-5-phosphate reductoisomerase]] can be inhibited by the antibiotic [[fosmidomycin]] which has been shown to potentially useful as an antimalarial.<ref name=Umeda2011>Umeda T, Tanaka N, Kusakabe Y, Nakanishi M, Kitade Y, Nakamura KT (2011) Molecular basis of fosmidomycin's action on the human malaria parasite ''Plasmodium falciparum''. Sci Rep 1:9</ref> |
|||
[[Isoprenoid]] biosynthesis may be inhibited by [[fosmidomycin]] which in turn reduces protein prenylation. One of the proteins that is normally prenylated is [[Rab5]] - a protein involved in vesicle transport. On inhibition by fosmidomycin the Rab5 proteins mislocalize and cause marked defects in food vacuolar morphology and integrity.<ref name=Howe2012>Howe R, Kelly M, Jimah J, Hodge D, Odom AR (2012) Isoprenoid biosynthesis inhibition disrupts Rab5 localization and food vacuolar integrity in ''Plasmodium falciparum''. Eukaryot Cell</ref> |
|||
[[HMB-PP synthase]] (IspG), an iron-sulphur (4Fe4S) protein involved in [[isoprenoid]] biosynthesis, has two domains - a TIM barrel and a 4Fe4S domain - in [[bacteria]]. In [[plant]]s and malaria parasites, there is an additional large insert domain.<ref name=Liu2012>Liu YL, Guerra F, Wang K, Wang W, Li J, Huang C, Zhu W, Houlihan K, Li Z, Zhang Y, Nair SK, Oldfield E (2012) Structure, function and inhibition of the two- and three-domain 4Fe-4S IspG proteins. Proc Natl Acad Sci USA</ref> This is a second TIM barrel that interacts with the other TIM barrel. |
|||
Rather than using [[sphingomyelin]] as the primary complex [[sphingolipid]], [[yeast]], [[plant]]s and some protozoa utilise an evolutionarily related [[inositol phosphorylceramide synthase]] to synthesize [[inositol phosphorylceramide]]. In the ''P. falciparum'' genome there is a single copy of a putative [[sphingomyelin synthase]].<ref name=Pratt2012>Pratt S, Wansadhipathi-Kannangara NK, Bruce CR, Mina JG, Shams-Eldin H, Casas J, Hanada K, Schwarz RT, Sonda S, Denny PW (2012) Sphingolipid synthesis and scavenging in the intracellular apicomplexan parasite, ''[[Toxoplasma gondii]]''. Mol Biochem Parasitol pii: S0166-6851(12)00283-6. {{DOI|10.1016/j.molbiopara.2012.11.007}}</ref> This gene's homolog in ''[[Toxoplasma gondii]]'' is the functional orthologue of the yeast's inositol phosphorylceramide synthase. |
|||
The parasite's requirement for [[acetyl-CoA]] are supplied by several pathways including [[acetyl-CoA synthetase]] and a [[pyruvate dehydrogenase]] like enzyme.<ref name=Cobbold2013>Cobbold SA, Vaughan AM, Lewis IA, Painter HJ, Camargo N, Perlman DH, Fishbaugher M, Healer J, Cowman AF, Kappe SH, Llinás M (2013) Kinetic flux profiling elucidates two independent acetyl-CoA biosynthetic pathways in ''Plasmodium falciparum''. J Biol Chem </ref> The nature of the pyruvate dehydrogenase like enzyme is not quite clear but it seems likely that the [[branched chain ketoacid dehydrogenase]] complex is performs this function. This latter pathway contributes glucose derived acetyl-CoA to the tricarboxylic acid cycle in a stage independent process whereas anapleurotic carbon enters the this cycle via a stage dependent [[phosphoenolpyruvate carboxylase]] / [[phosphoenolpyruvate carboxy-kinase]] process that decreases as the parasite matures. |
|||
;Membrane biogenesis |
|||
Membrane biogenesis in this organism involves the enzyme [[phosphoethanolamine methyltransferase]] which catalyses the methylation of [[phosphoethanolamine]] to [[phosphocholine]]. This pathway is found in [[plant]]s and [[nematode]]s but not in humans. The enzymes in ''P. falciparum'' is a multi-functional unlike that of plants and nematodes.<ref name=Lee2011>{{cite journal |author=Lee SG, Kim Y, Alpert TD, Nagata A, Jez JM |year=2011 |title=Structure and reaction mechanism of phosphoethanolamine methyltransferase from the malaria parasite ''Plasmodium falciparum'': An anti-parasitic drug target |journal=J Biol Chem}}</ref> The enzyme from ''P. falciparum'' has been cloned and its structure solved. |
|||
Four putative [[autophagy]] genes - those involved in ATG8 lipidation - are present in the genome.<ref name=Walker2013>Walker DM, Mahfooz N, Kemme KA, Patel VC, Spangler M, Drew ME (2013) ''Plasmodium falciparum'' erythrocytic stage parasites require the putative autophagy protein PfAtg7 for normal growth" ''PLoS One'' 8(6) e67047</ref> [[Atg8]] lipidation requires [[Atg7]] (an E1-type ligase), [[Atg3]] (an E2-type ligase) and [[Atg4]] (a cysteine protease). PfAtg7 is the activating enzyme of PfAtg8. PfATg7 appears to be essential for normal growth. |
|||
===Trace metals=== |
|||
Zinc is essential to the parasite and it actively accumulates it during growth.<ref name=Ginsburg1986>{{cite journal |last1=Ginsburg |first1=H |last2=Gorodetsky |first2=R |last3=Krugliak |first3=M |year=1986 |title=The status of zinc in malaria (Plasmodium falciparum) infected human red blood cells: stage dependent accumulation, compartmentation and effect of dipicolinate |journal=Biochim Biophys Acta |volume=886 |issue=3 |pages=337–344 |doi=10.1016/0167-4889(86)90168-0 |pmid=3518809}}</ref> During the life cycle zinc pools are formed in both the cytosol and the mitochondria.<ref name=Niles2012>Niles JC (2012) Malarial parasites accumulate labile zinc pools. Chem Biol 19(6) 660-661</ref> |
|||
A copper channel has been identified in the genome.<ref name=Asahi2013>Asahi H, Tolba ME, Tanabe M, Ohmae H (2013) Molecular factors that are associated with early developmental arrest of intraerythrocytic ''Plasmodium falciparum''. Can J Microbiol 59(7) 485-93. {{DOI|10.1139/cjm-2013-0166}}</ref> Removal of copper from the growth medium is associated with early developmental arrest. |
|||
===Vitamin A=== |
|||
The parasite takes up [[vitamin A]] from its host and uses it in own metabolism. Children with malaria have lower vitamin A levels than normal.<ref name=Mawson2013>Mawson AR (2013) The pathogenesis of malaria: a new perspective. Pathog Glob Health 107(3) 122-129</ref> |
|||
===Miscellaneous proteins=== |
|||
Polyamine biosynthesis in these parasites is controlled by a unique bifunctional [[S-adenosylmethionine decarboxylase]]/[[ornithine decarboxylase]] (PfAdoMetDC/ODC).<ref name=Williams2011>{{cite journal |author=Williams M, Sprenger J, Human E, Al-Karadaghi S, Persson L, Louw AI, Birkholtz LM |year=2011 |title=Biochemical characterisation and novel classification of monofunctional S-adenosylmethionine decarboxylase of ''Plasmodium falciparum'' |journal=Mol Biochem Parasitol |doi=10.1016/j.molbiopara.2011.07.004 |volume=180 |pages=17–26 |pmid=21803076 |issue=1}}</ref> On the secondary structure level PfAdoMetDC is similar to that of the human protein. This bifunctional enzyme ensure coordination decarboxylated AdoMet and [[putrescine]] for the subsequent synthesis of [[spermidine]]. |
|||
The first two reactions of the [[pentose phosphate pathway]] in ''P. falciparum'' are catalysed by a single bifunctional enzyme - [[glucose 6-phosphate dehydrogenase]] [[6-phosphogluconolactonase]].<ref name="Jortzik2011">{{cite journal |author=Jortzik E, Mailu BM, Preuss J, Fischer M, Bode L, Rahlfs S, Becker K |year=2011 |title=Glucose 6-phosphate dehydrogenase 6-phosphogluconolactonase: a unique bifunctional enzyme from ''Plasmodium falciparum'' |journal=Biochem J. |doi=10.1042/BJ20110170 |volume=436 |issue=3 |pages=641–50 |pmid=21443518}}</ref> This is distinct from the case in humans where the enzymes are separate. In [[animal]]s this pathway is usually found in the cytosol while in [[plant]]s it is found in the [[plastid]]s. The location of this reaction is not currently known in ''P. falciparum''. |
|||
Fusions between these two enzymes ([[glucose 6-phosphate dehydrogenase]] and [[6-phosphogluconolactonase]]) have also been reported in chordates.<ref name=Stover2011>{{cite journal |last1=Stover |first1=NA |last2=Dixon |first2=TA |last3=Cavalcanti |first3=AR |year=2011 |title=Multiple independent fusions of glucose-6-phosphate dehydrogenase with enzymes in the pentose phosphate pathway |journal=PLoS ONE |volume=6 |issue=8 |page=e22269 |doi=10.1371/journal.pone.0022269 |editor1-last=Moreno |editor1-first=Silvia N}}</ref> The chordate fusion differs in its orientation to that in ''Plasmodium'' (in ''Plasmodium'' the 6-phosphogluconolactone is found at the N-terminus of the glucose 6-phosphate dehydrogenase protein) indicating that at least two separate fusion events have occurred. The metazoan fusion appears to have occurred near the bases of the [[metazoan]] and [[apicomplexa]]n lineages. This fusion event was not found in any of the three sequenced ''[[Cryptosporidium]]'' genomes. It was not found in ''[[Perkinsus marinus]]'' or in either of the ciliate (''[[Paramecium tetraurelia]]'' and ''[[Tetrahymena thermophila]]'') genomes. More data will be needed to estimate the timing of this fusion event. |
|||
Only one of the two metazoan [[paralogs]] of glucose 6-phosphate dehydrogenase is fused, indicating that the fusion occurred after a duplication event. This duplication event occurred in an ancestor of the [[choanoflagellate]]s and metazoans. Another fusion event between these enzymes occurred in an ancestor of the protozoan parasites ''[[Trichomonas]]'' and ''[[Giardia lamblia]]''. In ''Giardia'', the proteins are fused in opposite orientations. A third fusion event occurred between glucose 6-phosphate dehydrogenase with [[phosphogluconate dehydrogenase]] in a [[diatom]] species (''[[Phaeodactylum tricornutum]]''). |
|||
The mechanism of action of the [[triose phosphate isomerase]] enzyme has been investigated in some detail.<ref name=Samanta2011>Samanta M, Murthy MR, Balaram H, Balaram P (2011) Revisiting the mechanism of the triose-phosphate isomerase reaction: The role of the fully conserved glutamic acid 97 Residue. Chembiochem {{doi|10.1002/cbic.201100116}}</ref> The conserved [[glutamic acid]] residue at position 97 is involved in the catalytic proton transfer. Modification of this residue may reduce the rate of catalysis by 9000 fold. |
|||
The [[shikimate]] pathways is functional in ''P. falciparum'' and [[vitamin E]] biosynthesis also occurs.<ref name=Sussmann2011>{{cite journal |author=Sussmann RA, Angeli CB, Peres VJ, Kimura EA, Katzin AM |year=2011 |title=Intraerythrocytic stages of ''Plasmodium falciparum'' biosynthesize vitamin E |journal=FEBS Lett |doi=10.1016/j.febslet.2011.11.005 |volume=585 |issue=24 |pages=3985–91 |pmid=22085796}}</ref> |
|||
The parasite actively synthesises [[pyridoxal-phosphate]] (vitamin B<sub>6</sub>). The [[2-C-methyl-d-erythritol-4-phosphate]] pathway is involved in its synthesis. This process involves two sets of reactions: condensation of [[ribulose 5-phosphate]], [[glyceraldehyde-3-phosphate]] and [[ammonia]] produced from [[glutamine]]. These actions are carried out by separate subunits. The synthase domain is known as Pdx1 and the [[glutaminase]] domain as Pdx2. In ''P. falciparum'' the core Pdx1 is a dodecamer and forms the core of the enzyme. There are up to 12 Pdx2 subunits surrounding the Pdx1 subunit.<ref>{{cite journal |author=Guédez G, Hipp K, Windeisen V, Derrer B, Gengenbacher M, Böttcher B, Sinning I, Kappes B, Tews I ''et al.'' |year=2012 |title=Assembly of the eukaryotic PLP-synthase complex from ''Plasmodium'' and activation of the Pdx1 enzyme |journal=Structure |volume=20 |issue=1 |pages=172–184 |doi=10.1016/j.str.2011.11.015 |pmid=22244765}}</ref> The majority of the synthesis is carried out by Pdx1. The pentose substrate is covalently attached through its C1 and forms a [[Schiff base]] with the Lysine 84 residue. The ammonia transfer between Pdx2 glutaminase and Pdx1 active sites is regulated by a transient tunnel. |
|||
[[Chorismate synthase]] (CS) catalyses the seventh and final step of the [[shikimate]] pathway. ''P. falciparum'' chorismate synthase (PfCS) is unique in terms of enzymatic behavior, cellular localization and in having two additional amino acid inserts compared to any other CS.<ref name=Tapas2011>{{cite journal |author=Tapas S, Kumar A, Dhindwal S, Preeti, Kumar P |year=2011 |title=Structural analysis of chorismate synthase from ''Plasmodium falciparum'': A novel target for antimalaria drug discovery |journal=Int J Biol Macromol |doi=10.1016/j.ijbiomac.2011.07.011 |volume=49 |issue=4 |pages=767–77 |pmid=21801743}}</ref> |
|||
There are several versions of the enzyme [[glutamate dehydrogenase]] (GDH) encoded in the genome. Of these, GDH1 and 3 appear to localise in the cytoplasm and GDH2 to the apicoplast.<ref name=Zocher2012>Zocher K, Fritz-Wolf K, Kehr S, Fischer M, Rahlfs S, Becker K (2012) Biochemical and structural characterization of ''Plasmodium falciparum'' glutamate dehydrogenase 2. Mol Biochem Parasitol</ref> |
|||
Within the genome, there are two [[adenylyl cyclase]]s - ACα and ACβ.<ref name=Salazar2012>Salazar E, Bank EM, Ramsey N, Hess KC, Deitsch KW, Levin LR, Buck J (2012) Characterization of ''Plasmodium falciparum'' adenylyl cyclase-β and its role in erythrocytic stage parasites" ''PLoS One'' 7(6) e39769.</ref> ACα contains six predicted transmembrane domains and a single carboxy-terminal catalytic domain homologous to sAC-like ACs. It is a predicted bifunctional protein comprising both a [[potassium]] channel and an AC that is conserved in the alveolates. It is expressed in the gametocytes. ACβ has no predicted transmembrane regions and possesses two AC catalytic domains. It has a marked [[pH]] dependence and is required for the erythrocytic stages. |
|||
Asymmetrical diadenosine 5',5″-P<sup>1</sup>,P<sup>4</sup>-tetraphosphate hydrolase (EC 3.6.1.17) catalyses the conversion of diadenosine 5',5″-P<sup>1</sup>,P<sup>4</sup>-tetraphosphate (Ap<sub>4</sub>A) to ATP and AMP and diadenosine 5',5″-P<sup>1</sup>,P<sup>5</sup>-pentaphosphate (Ap<sub>5</sub>A) to ATP and ADP. This enzyme from the parasite has been cloned and expressed.<ref name=Osman2012>Osman W, Endo S, Oh-Hashi K, Kitamura Y, Kitade Y (2012) Molecular characterization and mutational analysis of recombinant diadenosine 5',5″-P<sup>1</sup>,P<sup>4</sup>-tetraphosphate hydrolase from ''Plasmodium falciparum''. Biol Pharm Bull 35(7) 1191-1196</ref> |
|||
A [[glycerophosphodiesterase]] has been cloned.<ref name=Denloye2012>Denloye T, Dalal S, Klemba M (2012) Characterization of a glycerophosphodiesterase with an unusual tripartite distribution and an important role in the asexual blood stages of ''Plasmodium falciparum''. Mol Biochem Parasitol pii: S0166-6851(12)00222-8. {{DOI|10.1016/j.molbiopara.2012.09.004}}</ref> This enzyme is found in the parasitophorous vacuole, digestive vacuole and cytosol. It appears to be an essential gene but its specific function is currently unclear. |
|||
The [[translationally controlled tumor protein]] appears to bind [[artemisinin]].<ref name=Eichhorn2012>Eichhorn T, Winter D, Büchele B, Dirdjaja N, Frank M, Lehmann WD, Mertens R, Krauth-Siegel RL, Simmet T, Granzin J, Efferth T (2012) Molecular interaction of artemisinin with translationally controlled tumor protein (TCTP) of ''Plasmodium falciparum''. Biochem Pharmacol pii: S0006-2952(12)00678-8. {{DOI|10.1016/j.bcp.2012.10.006}}</ref> This may contribute to its anti malarial action. |
|||
A bifunctional [[farnesyl diphosphate synthase|farnesyl diphosphate]]/[[geranylgeranyl diphosphate]] synthase has been cloned.<ref name="Jordão2013">Jordão FM, Gabriel HB, Alves JM, Angeli CB, Bifano TD, Breda A, de Azevedo MF, Basso LA, Wunderlich G, Kimura EA, Katzin AM (2013) Cloning and characterization of bifunctional enzyme farnesyl iphosphate/geranylgeranyl diphosphate synthase from ''Plasmodium falciparum''. ''Malar J'' 12(1) 184</ref> It is encoded by the gene PF3D7_1128400. |
|||
[[Thiamine]] appears to be essential to the parasite's metabolism.<ref name=Chan2013>Chan XW, Wrenger C, Stahl K, Bergmann B, Winterberg M, Müller IB, Saliba KJ (2013) Chemical and genetic validation of thiamine utilization as an antimalarial drug target. Nat Commun 4:2060. {{DOI|10.1038/ncomms3060}}</ref> Thiamine is converted to its active form - thiamine pyrophosphate - by [[thiamine pyrophosphokinase]]. The enzymes [[oxoglutarate dehydrogenase]] and [[pyruvate dehydrogenase]] are both dependent on thiamine pyrophosphate. |
|||
A [[thymidylate kinase]] - an enzyme that catalyzes phosphorylation of thymidine monophosphate to thymidine diphosphate - is present in the genome.<ref name=Ojha2013>Ojha PK, Roy K (2013) First report on exploring structural requirements of alpha and beta thymidine analogs for PfTMPK inhibitory activity using in silico studies. Biosystems pii: S0303-2647(13)00165-2 {{DOI|10.1016/j.biosystems.2013.07.005}}</ref> |
|||
==Human immune system evasion== |
==Human immune system evasion== |
||
====''var'' family==== |
|||
The ''var'' genes encode for the ''P. falciparum'' erythrocyte membrane protein 1 ([[PfEMP1]]) proteins. The genes are found in the subtelomeric regions of the chromosomes. There exist an estimated 59 ''var'' genes within the genome.<ref name="gardner"/> |
|||
===''var'' family=== |
|||
The proteins encoded by the ''var'' genes are ultimately transported to the erythrocyte membrane and cause the infected erythrocytes to adhere to host endothelial receptors. Due to transcriptional switching between ''var'' genes, antigenic variation occurs which enables immune evasion by the parasite. |
|||
The ''var'' genes encode for the ''P. falciparum'' erythrocyte membrane protein 1 ([[PfEMP1]]) proteins. The genes are found in the subtelomeric regions of the chromosomes. There exist an estimated 59 ''var'' genes within the genome.<ref name="gardner"/> The proteins encoded by the ''var'' genes are ultimately transported to the erythrocyte membrane and cause the infected erythrocytes to adhere to host endothelial receptors. Due to transcriptional switching between ''var'' genes, antigenic variation occurs which enables immune evasion by the parasite. |
|||
====''rif'' family==== |
|||
The ''rif'' genes encode for repetitive interspersed family (rifin) proteins. The genes are found in the subtelomeric regions of the chromosomes. There exist an estimated 149 ''rif'' genes within the genome.<ref name="gardner"/> |
|||
The hypervariable ''var'' gene repertoire is to a large extent generated by frequent meiotic ectopic recombination in the mosquito gut. Mitotic recombination may also occur.<ref name=Duffy2009>Duffy MF, Byrne TJ, Carret C, Ivens A, Brown GV (2009) Ectopic recombination of a malaria var gene during mitosis associated with an altered ''var'' switch rate" ''J Mol Biol'' 389: 453–469 {{DOI|10.1016/j.jmb.2009.04.032}}</ref> |
|||
Rifin protein are ultimately transported to the erythrocyte membrane. The function of these proteins is currently unknown. |
|||
;Gene structure |
|||
====''stevor'' family==== |
|||
The ''stevor'' genes encode for the sub-telomeric variable open reading frame (stevor) proteins. The genes are found in the subtelomeric regions of the chromosomes. There exist an estimated 28 ''stevor'' genes within the genome.<ref name="gardner"/> |
|||
The ''var'' genes are classified into a number of subfamilies (A, B and C) that possess distinctive upstream and downstream flanking regions. A number of intermediate types - B/A and B/C - have also been described. The classification system is based primarily on the orientation of the genes: subtelomeric A and B family genes are oriented tail to tail (3′ to 3′) while central C family genes are oriented head to tail in a tandem repeat manner. The two intermediate groups - B/A and B/C- have chromosomal positions or domain composition that differ from those that would be expected from their orientation. |
|||
The function of the stevor proteins is currently unknown. |
|||
From N- to C-terminal the genes are organised in the following fashion: an N-terminal segment (NTS), Duffy binding-like (DBL) domains, Cysteine rich inter-domain regions (CIDR), C2 domains, one transmembrane region (TM) and the acidic terminal segment (ATS). |
|||
==Research== |
|||
{{seealso|Plasmodium molecular tools}} |
|||
The extracellular region of PfEMP1 comprises multiple adhesion domains called Duffy Binding Like (DBL) and Cysteine Rich Interdomain Region (CIDR). The number of DBL domains in each ''var'' gene varies between 2 and 9: the usual number is ~6. The DBL domains have been classified into different major classes - α, β, γ, δ, ζ and ε - and a number of sub-classes based on sequence criteria. Within the DBL are seven regions of considerable variability known as variable blocks (VB1-6). In all 147 subtypes have been recognised. In addition to these 21 conserved tandem runs of specific domains (domain cassettes) have also been identified.<ref name=Rask2010>Rask TS, Hansen DA, Theander TG, Gorm PA, Lavstsen T (2010) ''Plasmodium falciparum'' erythrocyte membrane protein 1 diversity in seven genomes--divide and conquer. PLOS Comput Biol 6: e1000933.</ref> The DBL domains have been also characterized by definition of 10 semi-conserved homology blocks (HBa-j) interspersed by hypervariable regions and by the definition of three structural subdomains (S1–3). Some biological correlations with the DBL domains have been described: DBLα has been associated with binding to [[heparin]] sulfate, [[blood group]] A antigen and [[complement receptor 1]]; DBLβ domains have affinity for [[ICAM-1]]; and DBLδ adheres to platelet-endothelial cell adhesion molecule 1 ([[PECAM-1]]).<ref name=Chen2000>Chen Q, Heddini A, Barragan A, Fernandez V, Pearce SF, ''et al''. (2000) The semiconserved head structure of ''Plasmodium falciparum'' erythrocyte membrane protein 1 mediates binding to multiple independent host receptors" ''J Exp Med'' 192: 1–10. {{DOI|10.1084/jem.192.1.1}}</ref> |
|||
==Resources== |
|||
[http://www.malaria.mr4.org/ MR4], The NIAID funded Malaria Research and Reference Reagent |
|||
Resource Center |
|||
The CIDR domains have been divided into three classes - α, β, and γ - and have three regions: the minimal CD36 binding region (M2) which is flanked flanked by the less conserved M1 and M3 regions.<ref name=Smith2000>Smith JD, Subramanian G, Gamain B, Baruch DI, Miller LH (2000) Classification of adhesive domains in the ''Plasmodium falciparum'' erythrocyte membrane protein 1 family. Mol Biochem Parasitol 110: 293–310. {{DOI|10.1016/S0166-6851(00)00279-6}}</ref> Several CIDRα class domains mediate binding to the human [[CD36 receptor]] but this binding only occurs in the B and C ''var'' families.<ref name=Robinson2003>Robinson BA, Welch TL, Smith JD (2003) Widespread functional specialization of ''Plasmodium falciparum'' erythrocyte membrane protein 1 family members to bind CD36 analysed across a parasite genome" ''Mol Microbiol'' 47: 1265–1278. {{DOI|10.1046/j.1365-2958.2003.03378.x}}</ref> The CIDRα domains also bind [[immunoglobulin M]] and PECAM-1.<ref name=Chen2000>Chen Q, Heddini A, Barragan A, Fernandez V, Pearce SF, ''et al'' (2000) The semiconserved head structure of ''Plasmodium falciparum'' erythrocyte membrane protein 1 mediates binding to multiple independent host receptors" ''J Exp Med'' 192: 1–10. {{DOI|10.1084/jem.192.1.1}}</ref> |
|||
[http://www.plasmodb.org/plasmo/home.jsp PlasmoDB] |
|||
An association between the length of the gene and its sequence variation has been reported.<ref name=Buckee2012>{{cite journal |last1=Buckee |first1=CO |last2=Recker |first2=M |year=2012 |month=Apr |title=Evolution of the multi-domain structures of virulence genes in the human malaria parasite, ''Plasmodium falciparum'' |journal=PLoS Comput Biol |volume=8 |issue=4 |page=e1002451 |doi=10.1371/journal.pcbi.1002451 |editor1-last=Antia |editor1-first=Rustom}}</ref> Shorter genes tend to be more variable and long genes to be more conserved. This probably is the result of a trade off between optimizing within host fitness and minimizing between host immune selection pressure. |
|||
[http://malaria.ucsf.edu/comparison/ Malaria IDC Strain Comparison Database] |
|||
;Evolution |
|||
[http://malaria.ucsf.edu/ Malaria IDC Transcriptome Database] |
|||
These genes appear to have evolved before the separation ''Plasmodium falciparum'' and ''[[Plasmodium reichenowi]].<ref name=Zilversmit2013>Zilversmit MM, Chase EK, Chen DS, Awadalla P, Day KP, McVean G (2013) Hypervariable antigen genes in malaria have ancient roots. ''BMC Evol Biol'' 13(1) 110</ref> |
|||
[http://sites.huji.ac.il/malaria/ Malaria Parasite Metabolic Pathways] |
|||
;Gene expression |
|||
[http://apicyc.apidb.org/ ApiCyc] |
|||
The ''var'' genes undergo antigenic switching - only a single gene is expressed at a given time and the expression varies during the course of infection. This variation begins when the merozoites leave the liver and the mechanisms driving this process are not presently understood. A method of erasing the epigenetic memory which appears to be involved in this process has been described.<ref name=Fastman2012>{{cite journal |last1=Fastman |first1=Y |last2=Noble |first2=R |last3=Recker |first3=M |last4=Dzikowski |first4=R |year=2012 |title=Erasing the epigenetic memory and beginning to switch-The onset of antigenic switching of ''var'' genes in ''Plasmodium falciparum'' |journal=PLoS ONE |volume=7 |issue=3 |page=e34168 |doi=10.1371/journal.pone.0034168 |editor1-last=Templeton |editor1-first=Thomas J}}</ref> An analysis of the results showed that a subtype of ''var'' genes - the upsA ''vars'' - which are rarely expressed in culture systems but appear to be important in clinical infections, are activated early in the switching process. The switching rate appears to be a function of the gene structure rather than its chromosomal position or promoter. |
|||
The protein PfSir2 associates with promoter regions of silenced genes involved in antigenic variation.<ref name=Figueiredo2005>Figueiredo L, Scherf A (2005) ''Plasmodium'' telomeres and telomerase: the usual actors in an unusual scenario. ''Chromosome Res'' 13(5) 517-524</ref> |
|||
A histone 3 lysine 4 [[methyltransferase]] - PfSET10 - which localizes exclusively to the perinuclear active ''var'' gene expression site and is required to maintain the active ''var'' gene in a poised state during division for reactivation in daughter parasites.<ref name=Volz2012>Volz JC, Bártfai R, Petter M, Langer C, Josling GA, Tsuboi T, Schwach F, Baum J, Rayner JC, Stunnenberg HG, Duffy MF, Cowman AF (2012) PfSET10, a ''Plasmodium falciparum'' methyltransferase, maintains the active ''var'' gene in a poised state during parasite division. ''Cell Host Microbe'' 11(1) 7-18</ref> |
|||
The variant silencing SET gene is an analog of the ''[[Drosophila melanogaster]]'' ASH1.<ref name=Jiang2013>Jiang L, Mu J, Zhang Q, Ni T, Srinivasan P, Rayavara K, Yang W, Turner L, Lavstsen T, Theander TG, Peng W, Wei G, Jing Q, Wakabayashi Y, Bansal A, Luo Y, Ribeiro JM, Scherf A, Aravind L, Zhu J, Zhao K, Miller LH (2013) PfSETvs methylation of histone H3K36 represses virulence genes in ''Plasmodium falciparum''. Nature {{DOI|10.1038/nature12361}}</ref> It controls histone H3 lysine 36 trimethylation (H3K36me3) on the ''var'' genes. Knocking this gene out results in the transcription of virtually all the ''var'' genes in the genome and their subsequent translation and localization to the cell membrane. This protein is present along the entire gene body including the intronic promoter and the transcription start site. |
|||
The [[origin recognition complex]] 1 protein binds the telomeres via its N terminal and appears to play some role in gene regulation.<ref name=Deshmukh2012>Deshmukh AS, Srivastava S, Herrmann S, Gupta A, Mitra P, Gilberger TW, Dhar SK (2012) The role of N-terminus of ''Plasmodium falciparum'' ORC1 in telomeric localization and ''var'' gene silencing. Nucleic Acids Res</ref> |
|||
Within the regulatory region of the ''var'' genes there are a number of insulator like elements.<ref name=Avraham2012>Avraham I, Schreier J, Dzikowski R (2012) Insulator-like pairing elements regulate silencing and mutually exclusive expression in the malaria parasite ''Plasmodium falciparum''. Proc Natl Acad Sci USA</ref> These along with the strict pairing of the 5' promoter with the second promoter within the intron are essential for the normal regulation of the ''var'' genes. |
|||
There is a small upstream (5') open reading frame associated with the var2csa gene.<ref name=Bancells2013>Bancells C, Deitsch KW (2013) A molecular switch in the efficiency of translation reinitiation controls expression of var2csa, a gene implicated in pregnancy associated malaria. Mol Microbiol {{DOI|10.1111/mmi.12379}}</ref> This gene is only expressed in placental malaria. The product of the open reading frame interacts with the sequence surrounding the var2csa gene to control its transcription. |
|||
Expression of gene appears to be non random with a global activation hierarchy favouring short and highly diverse genes in central chromosomal location.<ref name=Noble2013>Noble R, Christodoulou Z, Kyes S, Pinches R, Newbold CI, Recker M (2013) The antigenic switching network of ''Plasmodium falciparum'' and its implications for the immuno-epidemiology of malaria. Elife 2:e01074. {{DOI|10.7554/eLife.01074}}</ref> Longer and more conserved genes are rarely activated. |
|||
;Molecular biology |
|||
The intracellular portion of the EMP1 protein binds to PFI1780w - a member of the ''Plasmodium'' helical interspersed sub-telomeric (PHIST) family.<ref name=Mayer2012>Mayer C, Slater L, Erat MC, Konrat R, Vakonakis I (2012) Structural analysis of the ''Plasmodium falciparum'' Erythrocyte membrane protein 1 (PfEMP1) intracellular domain reveals a conserved interactionepitope. J Biol Chem</ref> |
|||
A low level solution of the ~300 kiloDalton ectodomain of PfEMP1 has been reported.<ref name=Brown2012>Brown A, Turner L, Christoffersen S, Andrews KA, Szestak T, Zhao Y, Larsen S, Craig AG, Higgins MK (2013) Molecular architecture of a complex between an adhesion protein from the malaria parasite and intracellular adhesion molecule 1. J Biol Chem</ref> It is rigid, elongated and monomeric and interacts with [[ICAM-1|intercellular adhesion molecule]] 1 through the Duffy binding Lβ domain alone, forming a 1:1 complex. |
|||
The strength of adhesion between the EMP1 and chondroitin sulfate A is decreased at 40C compared with its binding at 37C.<ref name=Carvalho2013>Carvalho PA, Diez-Silva M, Chen H, Dao M, Suresh S (2013) Cytoadherence of erythrocytes invaded by ''Plasmodium falciparum'': Quantitative contact-probing of a human malaria receptor. Acta Biomater pii: S1742-7061(13)00034-2. {{DOI|10.1016/j.actbio.2013.01.019}}</ref> |
|||
The subdomain 3 which connects the h6 and h7 α-helices of PfEMP1-DBL1α appears to be important in the process of parasitized erythrocytes forming rosettes with uninfected erythrocytes.<ref name=Angeletti2012>Angeletti D, Albrecht L, Blomqvist K, Quintana Mdel P, Akhter T, Bächle SM, Sawyer A, Sandalova T, Achour A, Wahlgren M, Moll K (2012) ''Plasmodium falciparum'' rosetting epitopes converge in the SD3-loop of PfEMP1-DBL1α" ''PLoS One'' 7(12) e50758. {{DOI|10.1371/journal.pone.0050758}}</ref> |
|||
;Clinical notes |
|||
It is thought that associations may exist between different ''var'' types and the clinical syndromes seen in malaria. The VAR2CSA variants bind to glycosaminoglycan chondroitin-sulfate A and have been associated with placental malaria.<ref name=Marsh1989>Marsh K, Otoo L, Hayes RJ, Carson DC, Greenwood BM (1989) Antibodies to blood stage antigens of ''Plasmodium falciparum'' in rural Gambians and their relation to protection against infection" ''Trans R Soc Trop Med Hyg'' 83: 293–303</ref> Group A and specifically Domanin Casette type 8 have been associated with cerebral malaria in Benin.<ref name=Bertin2013>Bertin GI, Lavstsen T, Guillonneau F, Doritchamou J, Wang CW, Jespersen JS, Ezimegnon S, Fievet N, Alao MJ, Lalya F, Massougbodji A, Ndam NT, Theander TG, Deloron P (2013) Expression of the Domain Cassette 8 ''Plasmodium falciparum'' erythrocyte membrane protein 1 is associated with cerebral malaria in Benin" ''PLoS One'' 8(7) e68368. {{DOI|10.1371/journal.pone.0068368}}</ref> Severe childhood malaria is associated with expression of specific PfEMP1 subtypes containing domain cassettes 8 and 13. These subtypes bind the [[endothelial protein C receptor]] (EPRC) with their amino terminal cysteine rich interdomain region.<ref name=Turner2013>Turner L, Lavstsen T, Berger SS, Wang CW, Petersen JE, Avril M, Brazier AJ, Freeth J, Jespersen JS, Nielsen MA, Magistrado P, Lusingu J, Smith JD, Higgins MK, Theander TG (2013) Severe malaria is associated with parasite binding to endothelial protein C receptor. Nature {{DOI|10.1038/nature12216}}</ref> This binding interferes with [[protein C]] binding to EPCR. |
|||
A difference between homology blocks has been noted in the phenotypes of erythrocyte rosetting and impaired consciousness.<ref name=Rorick2013>Rorick MM, Rask TS, Baskerville EB, Day KP, Pascual M. |
|||
Homology blocks of ''Plasmodium falciparum'' ''var'' genes and clinically distinct forms of severe malaria in a local population. BMC Microbiol 13(1):244 </ref> The clinical significance - if any - of this finding is not presently clear. |
|||
===''rif'' family=== |
|||
The ''rif'' genes encode for repetitive interspersed family (rifin) proteins. The genes are found in the subtelomeric regions of the chromosomes. There exist an estimated 149 ''rif'' genes within the genome.<ref name="gardner"/> |
|||
The rifins are divided into 2 types (A and B) depending on the presence/absence of a 25 amino acid domain in the semiconserved domain.<ref name=Mwakalinga2012>Mwakalinga SB, Wang CW, Bengtsson DC, Turner L, Dinko B, Lusingu JP, Arnot DE, Sutherland CJ, Theander TG, Lavstsen T (2012) Expression of a type B RIFIN in ''Plasmodium falciparum'' merozoites and gametes. ''Malar J'' 11(1) 429</ref> One of the rifins (PF13_0006) is transcribed both in the sexual and asexual stages. It is present on both gametocytes and merozoites. |
|||
Rifin proteins are ultimately transported to the erythrocyte membrane. The function of these proteins is currently unknown. |
|||
===''stevor'' family=== |
|||
The ''stevor'' genes encode for the sub-telomeric variable open reading frame (stevor) proteins. The genes are found in the subtelomeric regions of the chromosomes. There exist an estimated 28 ''stevor'' genes within the genome.<ref name="gardner"/> |
|||
These proteins appear to affect membrane deformability.<ref name=Sanyal2011>{{cite journal |author=Sanyal S, Egée S, Bouyer G, Perrot S, Safeukui I, Bischoff E, Buffet P, Deitsch KW, Mercereau-Puijalon O, David PH, Templeton TJ, Lavazec C |year=2011 |title=''Plasmodium falciparum'' STEVOR proteins impact erythrocyte mechanical properties |journal=Blood}}</ref> |
|||
==References== |
==References== |
||
{{reflist}} |
{{reflist|30em}} |
||
==Additional material== |
|||
* {{cite journal |author=Jewett , Sibley |year=2003 |title=Aldolase forms a bridge between cell surface adhesins and the actin cytoskeleton in apicomplexan parasites |journal=Mol Cell |volume=11 |issue=4 |pages=885–94 |doi=10.1016/S1097-2765(03)00113-8 |pmid=12718875 |first2=L.David}} |
|||
* {{cite journal |author=Bergman |year=2003 |title=Myosin A tail domain interacting protein (MTIP) localizes to the inner membrane complex of ''Plasmodium'' sporozoites |journal=J Cell Sci |volume=116 |issue=1 |pages=39–49 |display-authors=1 |doi=10.1242/jcs.00194}} |
|||
* {{cite journal |author=Baum |year=2005 |title=Invasion by ''P. falciparum'' merozoites suggests a hierarchy of molecular interactions |journal=PLoS Pathog |volume=1 |issue=4 |page=e37 |doi=10.1371/journal.ppat.0010037 |first2=Alexander G. |first3=Robert T. |first4=Ken M. |first5=Alan F.}} |
|||
* {{cite journal |author=Bosch |year=2007 |title=The closed MTIP-myosin A-tail complex from the malaria parasite invasion machinery |journal=J Mol Biol |volume=372 |issue=1 |pages=77–88 |display-authors=1 |doi=10.1016/j.jmb.2007.06.016 |pmid=17628590 |pmc=2702245 |first2=Stewart |first3=Claudia M. |first4=Thomas M. |first5=Lawrence W. |first6=Wim G.J.}} |
|||
* {{cite journal |author=Bosch |year=2007 |title=Aldolase provides an unusual binding site for thrombospondin-related anonymous protein in the invasion machinery of the malaria parasite |journal=Proc Natl Acad Sci USA |volume=104 |issue=17 |pages=7015–20 |doi=10.1073/pnas.0605301104 |pmid=17426153 |pmc=1855406 |first2=C. A. |first3=B. |first4=B. P. |first5=R. |first6=C. |first7=T. |first8=V. |first9=W. G. J.}} |
|||
* {{cite journal |author=Daher , Soldati-Favre |year=2009 |title=Mechanisms controlling glideosome function in apicomplexans |journal=Cur Opin Micro |volume=12 |issue=4 |pages=408–414 |doi=10.1016/j.mib.2009.06.008 |first2=Dominique}} |
|||
* {{cite journal |author=Sibley |year=2010 |title=How apicomplexan parasites move in and out of cells |journal=Cur Opin Biotech |volume=21 |issue=5 |pages=592–598 |doi=10.1016/j.copbio.2010.05.009}} |
|||
* {{cite journal |author=Hain and Bosch |year=2013 |title=Autophagy in Plasmodium, a multifunctional pathway? |journal=CSBJ |volume=8 |issue=11 |doi=10.5936/csbj.201308002 |first2=Jürgen}} |
|||
* {{cite journal |author=Boucher and Bosch |year=2013 |title=Development of a multifunctional tool for drug screening against plasmodial protein-protein interactions via surface plasmon resonance. | |journal=J. Mol. Recognit. |volume=26 |issue=10 |pages=496–500 |doi=10.1002/jmr.2292 |first2=Jürgen |pmid=23996492}} |
|||
==External links== |
|||
*[http://www.malaria.mr4.org/ MR4], The NIAID funded Malaria Research and Reference Reagent Resource Center |
|||
*[http://www.plasmodb.org/plasmo/home.jsp PlasmoDB] |
|||
*[http://www.genedb.org/Homepage/Pfalciparum GeneDB] |
|||
*[http://malaria.ucsf.edu/comparison/ Malaria IDC Strain Comparison Database] |
|||
*[http://malaria.ucsf.edu/ Malaria IDC Transcriptome Database] |
|||
*[http://sites.huji.ac.il/malaria/ Malaria Parasite Metabolic Pathways] |
|||
*[http://apicyc.apidb.org/ ApiCyc] |
|||
*[http://www.llamp.net/ Library of Apicomplexan metabolic pathways] |
|||
{{Alveolata}} |
|||
{{malaria}} |
|||
{{DEFAULTSORT:Plasmodium Falciparum Biology}} |
|||
[[Category:Plasmodium]] |
|||
[[ |
[[af:Plasmodium]] |
||
[[ar:متصورة]] |
|||
[[Category:Parasites]] |
|||
[[az:Malyariya paraziti]] |
|||
[[ca:Plasmodi]] |
|||
[[cs:Plasmodium]] |
|||
[[de:Plasmodium]] |
|||
[[et:Plasmoodium]] |
|||
[[es:Plasmodium]] |
|||
[[eo:Plasmodio]] |
|||
[[fr:Plasmodium]] |
|||
[[ko:말라리아원충]] |
|||
[[hi:प्लास्मोडियम]] |
|||
[[hr:Plasmodium]] |
|||
[[id:Plasmodium]] |
|||
[[it:Plasmodium]] |
|||
[[he:Plasmodium]] |
|||
[[ka:პლაზმოდიუმი]] |
|||
[[la:Plasmodium]] |
|||
[[lt:Plazmodis]] |
|||
[[ms:Plasmodium]] |
|||
[[nl:Plasmodium (eencellige)]] |
|||
[[pl:Zarodziec]] |
|||
[[pt:Plasmodium]] |
|||
[[ru:Плазмодии]] |
|||
[[sr:Plasmodium]] |
|||
[[sv:Plasmodium (släkte)]] |
|||
[[ta:பிளாஸ்மோடியம்]] |
|||
[[th:พลาสโมเดียม]] |
|||
[[uk:Плазмодій]] |
|||
[[vi:Ký sinh trùng sốt rét]] |
|||
[[zh:瘧原蟲]] |
|||
Revision as of 13:19, 12 November 2013
| Plasmodium falciparum | |
|---|---|
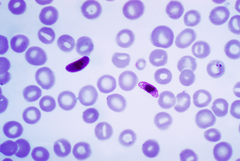
| |
| Blood smear with Plasmodium falciparum | |
| Scientific classification | |
| Domain: | |
| Kingdom: | |
| Superphylum: | |
| Phylum: | |
| Class: | |
| Order: | |
| Family: | |
| Genus: | |
| Species: | P. falciparum
|
| Binomial name | |
| Plasmodium falciparum Welch, 1897
| |
This article may be too long to read and navigate comfortably. (December 2012) |
Plasmodium falciparum has been the focus of much research due to it being the causative agent of malaria. This article describes some of the recent findings surrounding the unique biology of this organism.
Overview of life cycle
Plasmodium falciparum has a complicated life-cycle, requiring both a human and a mosquito host, and differentiating multiple times during its transmission/infection process.[1]

Genome
The genome of Plasmodium falciparum (clone 3D7) was fully sequenced in 2002.[3] The parasite has a 23 megabase genome, divided into 14 chromosomes.[3] The genome codes for approximately 5,300 genes. About 60% of the putative proteins have little or no similarity to proteins in other organisms and thus currently have no functional assignment.[3] It is estimated 52.6% of the genome is a coding region, with 53.9% of the putative genes containing at least one intron.[3]
Haploid/diploid
It is haploid during nearly all stages of its life-cycle, except for a brief period after fertilization when it is diploid from the ookinete to sporogenic stages within the mosquito gut.
AT richness
The P. falciparum genome has an AT content of roughly 80.6%.[3] Within the intron and intergenic regions, this AT composition rises to roughly 90%. The putative exons contain an AT content of 76.3%. The parasite's AT content is very high in comparison to other organisms. For example, the genomes of Saccharomyces cerevisiae and Arabidopsis thaliana are considered AT rich but have AT contents of 62% and 65%, respectively.[3]
Promoters
Although promoters are present in the genome, very little is known about them.
Recombination
The overall recombination rate is 9.6 kilobase per centimorgan and 54 candidate recombination hotspots have been identified.[4] The centromeres are found in chromosome regions largely devoid of recombination activity like other organisms. Within the hotspots a number of motifs were enriched including a 12 base pair G/C-rich motif with 3 base pair periodicity that may interact with a protein containing 11 predicted zinc finger arrays.
Subtelomeric regions
Throughout the eukaryotic kingdom, the overall structure of chromosome ends is conserved and is characterized by the telomeric tract - a series of short G-rich repeats. This is succeeded by an extensive subtelomeric region consisting of various types and lengths of repeats — the telomere associated sequences (TAS).[5] In general transcription of genes located next to telomeres is repressed, a phenomenon termed the telomere position effect. This effect is somewhat misnamed as it appears to be due to the sequences found in this region rather than the position of the gene.[6]
Subtelomeric regions in general are low in gene density, low in transcription, low in recombination, late replicating, are involved in protecting the end from degradation and end-to-end fusions and in completing replication. The subtelomeric repeats can rescue chromosome ends when telomerase fails, buffer subtelomerically located genes against transcriptional silencing and protect the genome from deleterious rearrangements due to ectopic recombination. They may also be involved in fillers for increasing chromosome size to some minimum threshold level necessary for chromosome stability; act as barriers against transcriptional silencing; provide a location for the adaptive amplification of genes; and be involved in secondary mechanism of telomere maintenance via recombination when telomerase activity is absent. The repressive histone 3 lysine 9 tri-methylation mark and heterochromatin protein 1 are found throughout the TAS region and adjacent gene families on all chromosomes. These heterochromatic marks are important in telomere proximal gene silencing.
In parasitic species genes involved in antigenic variation are commonly located in these regions.[7]
The chromsomes of P. falciparum conform to this basic pattern with the ends of the chromosomes consist of a stretch of telomeric GGGTT(T/C)A repeats with an average size of 1.2 kilobases (kb). This is followed by an extensive 20 to 40 kb TAS domain. These show a high degree of conservation within the genome and contain significant amounts of repeated structure.[3] Telomere repeats are followed by a mosaic of six distinct telomere associated repetitive elements (TAREs 1-6) which are always found in the same order but vary in size.
Many genes involved in antigenic variation are located in the subtelomeric regions of the chromosomes. These are divided into the var, rif, and stevor families. Within the genome, there are 59 var, 149 rif, and 28 stevor genes along with multiple pseudogenes and truncations.[3]
The chromosome ends form clusters of 4–7 telomeres that localize around the nuclear periphery.[8] Within this location the telomeric areas undergo frequent recombination which seems to increases antigenic variation.
Transcriptome

Transcription in P. falciparum appears to have significant differences from that found in most other eukaryotes examined to date with the chromatin undergoing dramatic unheavals during the cell cycle.[10]
A transcriptome analysis has been conducted on the intraerythrocytic development cycle of P. falciparum.[9] Roughly 60% of the genome is transcriptionally active during this portion of the parasite's life cycle. Whereas many genes appear to have stable mRNA levels throughout the cycle, many of the genes are transcriptionally regulated in a continuous cascade.
The transition from early trophozoite to trophozoite to schizont correlates with the ordered induction of genes related to transcription/translation machinery, metabolic synthesis, energy metabolism, DNA replication, protein degradation, plastid functions, merozoite invasion, and motility. Closely adjacent genes along the chromosome do not exhibit common transcription characteristics. Thus, genes are likely individually regulated along the parasite chromosome. Conversely, the apicoplast genome is polycistronic and most of its genes are coexpressed during the intraerythrocytic development cycle.[9]
Introns
The intron splicing has been examined experimentally.[11] The 5' and 3' splice sites agree with the canonical sequences (GT and AG respectively). The 5' consensus motif is weakly conserved and tolerates nucleotide substitution including the fifth nucleotide in the intron. This fifth position, typically a G nucleotide in most eukaryotes, is frequently an A in P. falciparum. The 3' splice site has a strong eukaryotic consensus sequence and a conserved adjacent polypyrimidine tract. The branch point is less well conserved with multiple branch points per intron with some at U instead of the typical A residue. A weak branch point consensus has been identified.
Proteome
There are 5,268 predicted proteins in Plasmodium falciparum and roughly 60% share little or no similarity to proteins in other organisms and thus are without functional assignment.[3] Of the predicted proteins, 31% contain at least one transmembrane domain and 17.3% have a signal peptide or signal anchor.[3]
It is estimated that 10.4% of the proteome is targeted to the apicoplast and 4.7% to the mitochondria.[3]
The parasite has different subsets of its proteome expressed during various stages of its developmental cycle.[12] In one study, of the 2,415 proteins were identified in four stages(sporozoite, merozoite, trophozoite, gametocyte), representing 46% of the theoretical number of proteins.[12] Only 6% of the proteins were found in all of the four stages. Of the proteins found, 51% were annotated as hypothetical proteins.
Merozoites contained high levels of cell recognition and invasion proteins. Trophozoites contained proteins implicated in erythrocyte remodeling and hemoglobin digestion. Gametocytes contained high amounts of gametocyte-specific transcription factors and cell cycle/DNA processing proteins. The gametocytes had low levels of polymorphic surface antigens. Sporozoites contained large amounts of proteins related to invasion, as well as members of the var and rif families.[12]
Translation initiation
This has been examined experimentally for the heat shock protein 86.[13] Like other eukaryotes purines at the -3 and +4 positions are essential for efficient translation. Uracil at the -1 position resulted in 2.5-fold higher reporter activity compared to wild type.
Mosquito bite
P. falciparum is transmitted to humans by the females of the Anopheles species of mosquito. There are about 460 species of Anopheles mosquito, but only 68 transmit malaria. Anopheles gambiae, found in Africa, is one of the best malaria vectors. It is long-lived, prefers feeding on humans, and lives in areas near human habitation.[14]
Prior to transmission, Plasmodium falciparum resides within the salivary gland of the mosquito. The parasite is in its sporozoite stage at this point. The Pumilio-FBF family member Puf2 appears to be critical for its survival in the mosquito salivary gland.[15]
As the mosquito takes its blood meal, it injects a small amount of saliva into the skin wound. The saliva contains antihemostatic and anti-inflammatory enzymes that disrupt the clotting process and inhibit the pain reaction.[16] Some of the details of this process are known. A salivary protein - anophelin - is a powerful thrombin inhibitor. This protein occupies thrombin's active site but is highly resistant to cleavage by the enzyme.[17]
Typically, each infected bite contains 5-200 sporozoites which proceed to infect the human vector.[14] Once in the human bloodstream, the sporozoites only circulate for a matter of minutes before infecting liver cells.
~1900 proteins have been reported from sporozoites infecting the salivary glands.[15]
Liver stage
After circulating in the bloodstream, the P. falciparum sporozoites enter hepatocytes. Before entering a hepatocyte the sporozoite typically engages in traversal of several cells. The reason for this behavior is not clear but it appears to reduce clearance of the sporozoites by Kupffer cells.[18]
Once within a hepatocyte the parasite loses its apical complex and surface coat and transforms into a trophozoite. Within the parasitophorous vacuole of the hepatocyte, P. falciparum undergoes schizogonic development. In this stage, the nucleus divides multiple times with a concomitant increase in cell size but without cell segmentation. This exoerythrocytic schizogony stage of P. falciparum has a minimum duration of roughly 5.5 days. After segmentation, the parasite cells differentiate into merozoites.[19]
Invasion of the hepatocytes appears to involve at least 2 proteins: sporozoite invasion-associated proteins (SIAP)-1 and -2.[20] These proteins bind heparin sulfate and chondroitin sulfate type membrane receptors on host cells.
Productive invasion of the hepatocyte results in the creation of a digestive vacuole, while merely passing through to reach another hepatocyte does not. Invasion of the liver cell changes the properties of the cell itself. The cell membrane becomes rougher and the cell itself becomes significantly stiffer.[21] The mechanism of these changes is currently unknown.
During this stage of development the sporozoite selectively discards organelles unnecessary for growth at this stage of the life cycle. Among these are the micronemes and the inner membrane complex.[22]
The division of the liver stages into thousands of merozoites is a complex process. In parallel with nuclear division, the apicoplast and mitochondrion become two extensively branched and intertwining structures.[23] The organelles subsequently undergo morphological and positional changes prior to cell division. Finally to form merozoites, the parasite undergoes cytokinesis.
After maturation, the merozoites are released from the hepatocytes and enter the erythrocytic portion of their life-cycle. Note that these cells do not reinfect hepatocytes.
Molecular biology of liver stages
Plasmodium possess only a single pyruvate dehydrogenase enzyme (PDH) complex. This is localized to the plastid-like organelle known as the apicoplast. Unlike most eukaryotes, Plasmodium lacks a mitochondrial PDH. The PDH catalyzes the conversion of pyruvate to acetyl-CoA, an important precursor for the tricarboxylic acid cycle and type II fatty acid synthesis.
The process of maturation within the liver is still being investigated. In the species Plasmodium yoelli the exit from the liver appears to be dependent on type II fatty acid synthesis.[24] Deletions in either the pyruvate dehydrogenase E1 alpha and E3 subunits produce a phenotype similar to that found in mutants of the type II fatty acid synthesis pathway. These mutants appear normal in blood stage development, mosquito stage development and early liver stage development but fail to exit the liver cells.
Plasmodium is unable to synthesize sterols they must obtain these from the host. However manipulation of cholesterol metabolism does not impede the development of the merozoites.[25]
Invasion of the hepatocyte induces the production of CD81 - a member of the tetraspanin superfamily.[26] CD81 also appears to play a role in liver invasion by Plasmodium species.[27] It is required for Plasmodium vivax sporozoite entry into human hepatocytes and for Plasmodium yoelii sporozoite entry into murine hepatocytes.[28]
The protein UIS3 is an essential protein for liver stage development.[29] It is thought to be localised to the membrane of the parasitophorous vacuole of the infected cell.
Latency of sporozoites is controlled by the eIF2 alpha kinase IK2, a general inhibitor of protein synthesis.[30] Puf2 participates in the regulation of IK2 and inhibits premature sporozoite transformation. In contrast Puf1 appears to be dispensable.
The RNA binding protein family PUF member Pumilio-2 (Puf2) appears to be involved in transformation of sporozoites into the hepatic stage of the life cycle.[31] Knock out mutants of this gene exhibit genome wide transcriptional changes resulting in loss of gliding motility, cell traversal ability, reduction in infectivity and trigger metamorphosis typical of early Plasmodium intra-hepatic development.
Type II fatty acid biosynthesis is vital for this stage in the life cycle.[32] This pathway may be inhibited by the antibiotic triclosan.
The host iron regulatory hormone hepcidin which is synthesised in the liver and spleen, appears to be able to inhibit growth of the liver stages.[33] Levels of this hormone are elevated during infection and seem to correlate with the anaemia often found in malaria.[34] Erythrocytic parasitaemia, above a minimum threshold, impairs the growth of subsequent liver cell sporozoite infection.[33] The production of hepcidin leads to the redistributes iron away from hepatocytes thus slowing the development of the iron dependent liver stage.
Liver hepcidin expression is upregulated and downregulated during the early and late stages of malaria infection respectively.[35] Inflammation and erythropoietin, rather than the iron sensing pathway, are involved in the regulation of hepcidin expression. Treatment of malaria infected mice with anti hepcidin neutralizing antibodies increased parasitemia and mortality rates. Overexpression of hepcidin improves the outcome.
Lipocalin 2, a host protein that sequesters iron, is upregulated during infection and appears to be involved in the host response.[36] This protein increases both host macrophage function and granulocyte recruitment and decreases reticulocytosis.
Expression of the iron sequestering protein ferritin (ferritin H chain in mice) is associated with decreased tissue damage.[37] The mechanism appears to be via prevention of activation of the proapoptotic c-Jun N-terminal kinase.
Invasion of the hepatocyte seems to require the RON4 protease.[38]
Within the liver actin reorganization is a dynamic process in part controlled by the actin severing and capping protein - gelsolin.[39] The hepatocyte cytoskeleton may contribute to parasite elimination.
Within the genome is encoded a homolog of macrophage migration inhibitory factor. This gene appears to be important for parasite development in the liver.[40]
In Plasmodium bergei a protein - liver specific protein 2 (LISP2) - is expressed 24 hours after infection and rapidly increases during the liver stage schizogony. LISP2 is carried first to the parasitophorous vacuole and subsequently transported to the cytoplasm and nucleus of host hepatocytes. Mutations in this gene result in arrested development of the merozoites.[41]
Two other proteins (p52 and p36)in Plasmodium bergei appear to be important in the formation of the parasitophorous vacuole membrane in the liver.[42]
Infection of the liver induces apotosis in some of the liver cells.[43] Blocking this apototic response seems to increase the number of parasites in the liver suggesting that this may be a host defense mechanism.
Erythrocytic stage
Effects on erythrocyte
Infection of the erythrocyte induces a series of changes in the host cell's membrane. These changes depend on the stage of infection and are due to a subset of parasite derived proteins that are exported across the parasic vaculole membrane into the host cell's cytosol where they interact with the host cell cytoskeleton or are exposed at the erythrocyte surface.
Among these changes are a loss of deformability, an increase in rigidity and a novel propensity to adhere to vascular endothelial cells and unparasitized erythrocytes. Several proteins including knob-associated histidine rich protein, Plasmodium falciparum erythrocyte membrane protein 3, mature parasite infected erythrocyte surface antigen and ring parasite infected erythrocyte surface antigen are known to bind to the cytoskeleton and increase the erythrocyte's rigidity. Others including the Plasmodium falciparum erythrocyte membrane protein 1 - the product of the var gene - are located on the surface of the erythrocyte and cause and are responsible for its new tendency to adhere.
Another induced change is an alteration in the zeta potential - an electrochemical property of cell surfaces that is determined by the net electrical charge of molecules exposed at the surface of cell membranes - of the cell. The normal zeta potential of the erythrocyte is -15.7 millivolts (mV).[44] Much of this potential appears to be contributed by the exposed sialic acid residues in the membrane: their removal results in zeta potential of -6.06 mV. Infection of the erythrocyte decreases the mean to -14.6 mV at the trophozoite stage. Again removal of sialic acid in the infected cells increased the potential to -4.64 mV.
A further change is the appearance of knobs - localised swellings of the erythrocyte membrane. The number of knobs, their diameter and height vary between isolates. They are thought to be important in the pathogenesis by contributing to blockage of capillaries. A number of parasite proteins are associated with these knobs including erythrocyte membrane protein 1 (the var gene product) and knob-associated histadine rich protein. Knobs first appear ∼20 hour post invasion and increase in number to ∼35 hours post-invasion.[45] They are more common in ex vivo isolates than in culture maintained strains.
Singlet oxygen is generated during the parasite's life cycle. Its production is least in the ring stages, maximal in the schizonts and intermediate in the trophozoites.[46] Its production may be related to the formation of haemozoin.
In normal erythrocytes the proteins ankyrin, band 3, band 4.1, glycophorin and spectrin are phosphorylated. After invasion by the merozoite these proteins become dephosphorylated.[47] As the parasite matures selective phosphorylation of ankyrin, band 3 and spectrin occurs.
Tyrosine phosphorylation of band 3 at the ring stage appears to be under the control of Syk kinase.[48] Phosphorylation of additional cytoskeletal, trans-membrane and membrane associated proteins occurs as the parasite matures. These include actin, adducins, band 4.2 and catalase. During the late schizont stage widespread protein dephosphorylation occurs. The erythrocyte kinases may be involved in this process.
Aggregation of the erythrocytes is known to occur during infection. This effect can be caused by culture supernatant suggesting a soluble product is responsible. Part of this mechanism appears to be the externalization of phosphatidyl-serine residues in the erythrocyte membrane. Methaemoglobin has been identified as the main causative agent of this alteration.[49] Its mechanism of action appears to be via the generation of reactive oxygen species: this action may be reversed with the addition of antioxidants.
Metalloproteinase 9 is released from human microvascular endothelium after contact with infected erythrocytes.[50]
Significant changes occur in the erthrocyte's cytoskeleton during infection.[51] Among these are the accumulation of spectrin around the knobs and a decrease elsewhere.
The change in the shape of the erythrocyte induced by the parasite depends on the parasite species.[52] Infection with P falciparum induces an increase in the cell volume of 80% in the ring stage. The erythrocyte subsequently becomes spherical in the trophozoite stage and remains so in the schizont stage. In contrast the volume increase induced by P vivax is only 30% and the erythrocyte remains biconcave throughout the infective cycle.
Merozoite
After release from the hepatocytes, the merozoites enter the bloodstream prior to infecting red blood cells. At this point, the merozoites are roughly 1.5 µm in length and 1 µm in diameter and use the apicomplexan invasion organelles (apical complex, pellicle and surface coat) to recognize and enter the host erythrocyte.
The apicoplast measures 0.5 µm × 0.15 µm in the merozoite and is anchored to a band of 2-3 subpellicular microtubules.[53] Within the merozoite the mitochondrion and the apicoplast are aligned asymmetrically along the same side as the microtubules.
There are up to 40 micronemes per merozoite shaped like longnecked bottles. They are ~160 nanometers (nm) long and 65 nm at their widest diameter.[54] On their external surfaces, they bear bristle like filaments, each 3-4 nm thick and 25 nm long. The micronemes are translocated from a single Golgi like cisterna near the nucleus along a band of two or three subpellicular microtubules to the merozoite apex where they dock with the rhoptry tips.
Unlike species in the genus Toxoplasma which have multiple rhopteries, Plasmodium species typically only have two. These may be referred to in the literature as the 'paired organ'. Several proteins have been localised to the rhopteries including asparagine rich parasite protein encoded on chromosome 4.[55] Another is the Apical rhoptry neck protein whose expression is confined to the schizont stage.[56]
After release from the erythrocyte the merozoites of the rodent malaria parasite Plasmodium yoelii change their shape from flat elongated ovals to spherical bodies.[57] This process takes ~60 seconds. During this time the merozoites were able to attach to and deform the erythrocyte membrane but were not able to reorient and invade. This morphological change may be related to the secretion or activation of invasion related proteins.
The parasite first binds to the erythrocyte in a random orientation. It then reorients such that the apical complex is in proximity to the erythrocyte membrane. A tight junction is formed between the parasite and erythrocyte. As it enters the red blood cell, the parasite forms a parasitophorous vesicle, to allow for its development inside the erythrocyte.[58]
Five separate invasion paths have been delineated.[59] The most common pathway is neuraminidase resistant, trypsin sensitive and chymotrypsin resistant invasion. Some parasites have a neuraminidase- and trypsin-sensitive phenotype indicating a dependence on the erythrocyte binding antigen 175/glycophorin A pathway(s). Most isolates appear to be dependent on a trypsin sensitive pathway.
Erythrocyte invasion
This is a complex and poorly understood process. The merozoite initially contacts the erythrocyte and rotates until the rhoptery containing part is adjacent to the erythrocyte membrane. A tight contact is then established and the parasite enters the erythrocyte. This happens within seconds making the invasion process difficult to analyse. In Plasmodium yoelii the serine type serine repeat antigen (SERAs) are non essential for blood stage development of the parasite but appear to be an important factor in enabling the parasite to fully utilize the whole age repertoire of circulating erythrocytes.[60]
In culture invasion of the erythrocytes can be prevent with the use of heparin.[61] Heparin binds only at the apical tip of the merozoite surface and multiple heparin binding proteins are found preferentially in the apical organelles.[62] The Duffy and reticulocyte binding-like families bind to heparin with diverse affinities suggesting that heparin masks the apical surface of merozoites and blocks interaction with the erythrocyte membrane after initial attachment.
Another protein that is bound by heparin is the merozoite surface protein 1.[63]
262 open reading frames show sharp induction of expression during late schizont stages. At least 28 of these are proteins that are known to be involved in the invasion process. The functions of over 190 of these remain unknown.[64]
Initial adhesion
The rhopties are an ancient organelle within this group of protozoa while the micronemes appear to be a more recent development. The rhopteries are involved in cell invasion in other species in this clade. It would appear that the micronemal proteins have become adapted to recognise a cell potentially suitable for invasion and that the rhoptry proteins facilitate the invasion itself. It is likely that there is overlap of these functions particularly in the invasion step and that other proteins are also involved.
Exposure of the merozoites to a low potassium medium - at a concentration similar to that found in blood - induces a rise in the merozoite's cytosolic calcium concentration.[65] The low potassium levels in the blood (3.5-5.0 millimoles per liter) activates a phospholipase C enzyme.
The calcium dependent protein kinase 1 appears to play a role in micronene discharge.[66] The drug purfalcamine is a specific inhibitor of this kinase and it also inhibits micronene discharge and erythrocyte invasion. These kinases typically have an N terminal kinase domain and C terminal calmodulin like domain with calcium binding EF hands. The N and C terminals are joined by a junction domain. The C terminal appears to interact with the junction domain in the process of binding calcium.
Calcineurin a Ca2+ dependent protein phosphatase is also involved in this process.[67] The dephosphorylation and depolymerization of apical actin also seems to be involved in this process.
Phospholipase C initiates a pathway that leads to a rise in the intracellular calcium. This rise in calcium triggers secretion of microneme proteins including the 175 kD erythrocyte binding antigen and apical membrane antigen 1 to the merozoite surface where they can bind to erythrocyte surface proteins including glycophorin A. The interaction of EBA175 with glycophorin A, its receptor on erythrocytes, restores basal cytosolic calcium levels. This in turn triggers the release of rhoptry proteins including the thrombospondin related apical merozoite protein (TRAMP).[68]
GAP45 is phosphorylated in response to Phospholipase C and calcium signaling.[69] It is phosphorylated by the P. falciparum kinases Protein kinase B and Calcium dependent protein kinase 1, both of which are calcium dependent enzymes, at Serine89, Serine103 and Serine149. Phosphorylation of these sites is differentially regulated during parasite development.
Two families of proteins are known to be involved in the invasion process: the reticulocyte binding like homologues (PfRh or PfRBP) and erythrocyte binding like (EBL) proteins. The EBL family are principally located in the micronemes and the Reticulocyte binding Homolog (PfRH) family are principally located in the rhopteries. Ligands from the EBL family largely govern the sialic acid dependent pathways of invasion and the RH family ligands (except for RH1) mediate sialic acid independent invasion.[70] During the invasion process these ligands are localized at the apical tip of the merozoite and are able to bind erythrocytes.
- Microneme proteins
All known micronemal proteins are type I integral membrane proteins that contain a C-terminal transmembrane domain and a short cytoplasmic domain.[71]
The EBL family of proteins includes EBA-165 (also known as PEBL), EBA-175 (also known as PfF2), EBA-181 (also known as JESEBL), EBA-140 (also known as BAEBL) and EBL-1. Whilst these parasite ligands function in merozoite invasion by binding to specific receptors on the erythrocyte, they appear also to have a central role in activation of the invasion process. Binding of EBA-175 to its receptor, glycophorin A, restores the basal cytosolic calcium levels after interaction of the merozoite with the erythrocyte and triggers the release of rhoptry proteins.[65]
These proteins have several domains. Region II which is responsible for ligand erythrocyte interaction during invasion, consists of two homologous F1 and F2 domains.[72]
Several of the receptors for the proteins are known: glycophorin A for EBA-175, glycophorin B for EBL-1 and glycophorin C for EBA-140.[73][74][75][76]
The ebl family share a common intron/exon structure suggesting a common origin.[77] These proteins also share a common organisation into several regions. Of these regions II and VI are the cysteine-rich regions of the DBL-EBP extracellular domains that have numerous conserved cysteine and hydrophobic amino acid residues, suggesting a conserved, functionally important three-dimensional structure. Despite this there is little nucleotide identity between the proteins.
The EBL proteins have a Duffy like binding domain (DBL) unique to Plasmodium species. The crystal structure of the binding domain of EBA-175 has been solved.[78] It consists of a dimer of the beta fingers of the F1 and F2 regions.
The crystal structure of EBA-140 has been solved.[79] The two domain binding region is present as a monomer. Both domains are required for binding to occur. Its electrostatic surface has a basic patch spanning both DBL domains that is important in the binding mechanism.
EBA-175 mediates adhesion to erythrocytes through binding of the Duffy binding like domains in its extracellular domain to Neu5Acα2-3Galactose displayed on the O-linked glycans of glycophorin A.[80].
A member of the EBL family of proteins (MAEBL) has been shown to be present in Plasmodium gallinaceum.[81] This protein is now known to be conserved in the primate, rodent and avian infecting species suggesting that it may play an important role in erythrocyte invasion. The duplicate extracellular binding domains of MAEBL are responsible for erythrocyte binding. MAEBL is a type I transmembrane protein with a carboxyl cysteine rich region.
A GPI-anchored micronemal antigen (GAMA) also appears to be essential in the process of erythrocyte invasion.[82]
A microneme associated antigen (PfMA: PF3D7_0316000, PFC0700c) binds erythrocytes in a sialic acid independent, chymotrypsin and trypsin resistant manner.[56] This gene is expressed only in the late blood stages. It is a 307 amino acid protein (~ 37.1 kiloDalton) that contains an N-terminal stretch of hydrophobic residues, a C-terminal single transmembrane domain and a short cytoplasmic tail. It is moderately (~40%) conserved between several Plasmodium species.
- Rhoptery proteins
The PfRh family consists of five proteins and a pseudogene: PfRh1, PfRh2a, PfRh2b, PfRh3, PfRh4 and PfRh5. PfRh3 is a transcribed pseudogene in all strains examined to date.[83] All the other members of this family bind to erythrocyes and antibodies to them inhibit invasion.
The Pfh1 protein binds a sialic acid containing erythrocyte receptor.[84]
PfRh1 and PfRh2 are located in the neck of the rhopteries.[85][86]
The genes PfRh2a and PfRh2b encode large proteins of about 3200 amino acids in length. They differ only in the last 500 amino acids of the C terminal and have clearly arisen by a process of gene duplication and mutation. The 500 amino acid region includes an ectodomain, a transmembrane domain and a cytoplasmic domain. PfRh2b is essential for a well-defined invasion pathway while PfRh2a is not required or sufficient for this pathway. It has been shown that the reason for this difference lies in the cytoplasmic domain.[87]
Reticulocyte binding like protein homologue 2a (PfRH2a) is processed both in the schizont as well as during invasion resulting in proteins with different erythrocyte binding properties.[88] It also moves from the rhoptry neck to the moving junction during merozoite invasion. PfRh2a undergoes a cleavage event in the transmembrane region during invasion consistent with activity of the membrane associated PfROM4 protease.[89] Both PfRh2a and PfRh2b bind to red blood cells. The erythrocyte-binding domain lies within a 15 kDa region at the N-terminus of each protein.
PfRh2b appears to play an important - if not dominant - role in the binding and invasion process.[90]
PfRh4 binds to the complement receptor 1 (CR1; CD35).[91] Complement receptor 1 is a ∼190- to 280-kDa single-chain transmembrane glycoprotein and carries the Knops blood group antigen. The binding site lies within the three N-terminal complement control protein modules (CCPs 1-3) of CR1. This region also accommodate binding and regulatory sites for the key complement activation specific proteolytic products, C3b and C4b. The binding of Rh4 to CR1 does not inhibit the binding of C3b/C4b but it does inhibit their dissociation from the erythrocyte. The critical site for the binding of Rh4 appears to lie within the CCP-1 module.
PfRh4 is responsible for the majority (50–80%, depending on the parasite strain used) of the sialic acid independent invasion pathway.[92]
PfRh4 binds to a second protein P. falciparum Rh5 interacting protein (PfRipr). PfRipr has a molecular weight of 123 kiloDaltons with 10 epidermal growth factor-like domains and 87 cysteine residues distributed along the protein.[93]
PfRh5 is located within the rhoptries and appears to be an essential gene.[94]
The receptor for the PfRh5 protein appears to be the Ok blood group antigen, basigin.[95] Blocking access to this protein on the erythrocyte surface appears to inhibit erythrocyte invasion completely. Binding of the Rh5 protein appears to be critically dependent on a single residue within the Rh5 protein.[20] Antibodies to this protein inhibit the parasite's growth in vitro and appear to be correlated with protection against infection with it.[96]
There seems to be some overlap between the functions of these proteins. Loss of EBA-175 can be compensated by increased expression of PfRh4.[97]
The rhoptry associated, leucine zipper-like protein 1 (RALP1) is specifically expressed in schizont stages and localized to the rhoptry of merozoites. It appears to be an essential gene. It translocates from the rhoptry neck to the moving junction during merozoite invasion. Anti-RALP1 antibodies disrupt the tight junction formation. There is an erythrocyte binding domain in the C terminal.
Another protein localised to the apical ends of the rhoptries is an asparagine rich parasite protein (PfAARP;PFD1105w).[98] This protein has a predicted signal sequence, a C-terminal transmembrane region and its transcription and translation patterns are similar to other merozoite surface proteins.
The rhoptry neck protein PfRON4 - a homologue of Toxoplasma gondii rhoptry neck protein TgRON4 - forms a complex with the protein PfAMA1 during its secretion in the course of merozoite invasion.[99]
Other rhoptery proteins include the rhoptry associated protein 1 (RAP1) and rhoptry associated membrane antigen (RAMA).[100][101]
The RhopH complex proteins localize to the basal bulb of the rhoptries and are involved in erythrocyte binding and in establishment of parasitophorous vacuole.[102][103]
Apical membrane antigen 1 (AMA-1) is a type I transmembrane protein located in the neck of the malaria merozoite rhoptries and later on the surface of the invasive merozoite.[104] Its ectodomain is defined by three cysteine-rich domains characterised by disulfide bond patterns.
- Merozoite surface protein family.
Merozoite surface protein 1 (MSP-1; P195; PMMSA; MSA 1) is a protein found on the surface of the merozoites.[105] During invasion of the new red cell most of the MSP1 molecule is shed from the parasite surface except for a small C-terminal fragment which can be detected in ring stages. Within this fragment are two epidermal growth factor-like domains. This protein has been found in all Plasmodium species studied to date suggesting it has an important role in the life cycle.
It appears that the merozoite surface protein 1 (MSP1) binds to heparin like molecules on the surface of the erythrocyte and that is binding is an essential step in the invasion process.[63]
Merozoite surface protein 2 is one of the most abundant proteins on the surface of the merozoite and plays a role in the invasion process.[106] Mediated by hdrophobic residues in the N terminal 1-25 residues it forms aggregates in vitro and may be present on the merozoite as aggregates.
In P. vivax there are a family (10) of merozoite surface protein 3 termed MSP3α to MSP3λ arranged in a head to tail fashion on chromosome 10.[107] These proteins have a predominant central alanine rich domain containing heptad repeats predicted to form α-helical secondary and coiled-coil tertiary structures. They lack transmembrane domain or GPI-lipid modification site.
Merozoite surface protein 8 is a single open reading frame of 1791 base pairs which encodes a polypeptide of 597 amino acids.[108] At the N-terminus there is a secretory signal peptide while at the and C-termini there is a GPI attachment sites. There are two EGF-like domains located near the C-terminus.
Merozoite surface protein 9 is conserved between species and appears to be under purifing selection.[109]
Merozoite surface proteins 8 and 10 which are thought to be involved in the invasion process appear to be under purifying selection.[110]
- Other proteins
Another family of proteins involved in the invasion process are the thrombospondin related anonymous protein (TRAP) family. These proteins are type I cell surface proteins with one or more extracellular thrombospondin type-I repeats (TSR) domains and/or von Willebrand factor like (vWF) A domain(s) and an acidic cytoplasmic tail with a subterminal tryptophan residue. The cytoplasmic tails of TRAP, CTRP, TLP and MTRAP interact with the enzyme aldolase.
One protein thought to be involved in the invasion process is the merozoite specific thrombospondin related anonymous protein homolog (MTRAP). The receptor for this protein has been identified as the GPI-linked protein semaphorin-7A (CD108).[111][112] The MTRAP monomers interact via their tandem TSR domains with the Sema domains of a Semaphorin-7A homodimer.
The motile forms have their own stage specific cell surface TRAP family member: TRAP and S6 (also known as TREP) occur on the sporozoites; CTRP is found on the ookinetes; MTRAP is expressed in the merozoites; and TLP is present on both sporozoites and merozoites. Other members of this family are the proteins CSP, SPATR, TRSP, WARP and PTRAMP. Roles for several of these proteins has been discovered: TRAP is critical for sporozoite invasion of the mosquito salivary glands, infection of mammalian liver and sporozoite gliding motility; CTRP is required for invasion of the mosquito midgut; and S6 is important for both sporozoite gliding motility and invasion of mosquito salivary glands. TLP has a role in sporozoite cell traversal. The cytoplasmic tail of TRAP is essential for gliding motility and invasion of the mosquito's salivary glands. Both the TSR and A domains of TRAP are required for the invasion of the mosquito salivary glands. Penetration of the mammalian hepatocytes however requires the TSR, the A domain and the cytoplasmic tail. In contrast only the A domains of CTRP are essential for infectivity by the ookinete.
PfTRAMP is localised to the base of the rhopteries.[113]
The protein PfTCTP causes the release of histamine in the host.[114] This protein also activates the basophils.
The cysteine rich protective antigen appears to play a role in this process.[115]
The ribosomal phosphoprotein P0 also seems to be involved in the invasion process.[116]
A double C2 domain (DOC2) protein appears to be involved in the invasion of the erythrocyte.[117] DOC2 proteins recruit the membrane fusion machinery an essential part of the Ca2+-dependent exocytosis mechanism.[118] These proteins have a Munc13-interacting domain and tandem C2s (designated C2A and C2B) which are connected by a short polar linker. The C2 domains bind phospholipids in a Ca2+-dependent manner. Elucidating their precise role in erythrocyte invasion requires further work.
In Plasmodium vivax a number of tryptophan rich antigens are involved in erythrocyte invasion.[119] Homologs of these proteins are found in P. falciparum - tryptophan-threonine rich antigen (PfTryThrA) and merozoite associated tryptophan rich antigen (PfMaTrA) and Plasmodium yoelii. These proteins also seem to be involved in the invasion process.
In P. vivax the asparagine rich protein has been cloned.[120] It is a 281-residue-long molecule, which is encoded by a single exon and has an N-terminal secretion signal in addition to a tandem repeat region. This protein is expressed in mature schizonts and is located on the merozoite surface and appears to accumulate towards the apical pole.
Another protein that appears to be involved in the invasion process is PfDBLMSP (PF10_0348).[121] This protein has a predicted signal sequence, a central Duffy binding-like (DBL) domain and a secreted polymorphic antigen associated with merozoites (SPAM) domain in its C-terminal half. The transcription and translation of this gene is up regulated specifically in schizonts, similar to other merozoite proteins involved in invasion of erythrocytes. This protein seems to be under selection pressure.
Invasion
The process of invasion is partly understood. The merozoite proteins forms a tight junction (the moving junction complex) with some of the erythrocyte membrane proteins.[122] The attached merozoite proteins are then moved posteriorly by an actin-myosin motor. The net effect of this process is to drive the merozoite into erythrocyte.
Some details of the invasion process are known.[123] The rhoptery protein RON2 is inserted into the erythrocyte membrane. The protein AMA1 secreted from microneme then binds to RON2. RON2 forms part of a macromolecular complex which includes RON2, RON 4, RON5 and RON8. The protein PfRON2 via its C-terminal as well as its central cysteine rich domain interacts with PfAMA1.[124]
The membrane proximal domain of the AMA1 protein is responsible for direct binding to erythrocytes.[125]
The invasion process appears to be ATP dependent[126] and may involve a purogenic signalling pathway.
A protein that appears to be unique to the genus Plasmodium - RON12 has been described.[127] RON12 lacks membrane anchors and is a major soluble component of the nascent parasitophorous vacuole. Most of the secretion of RON12 occurs late during invasion (after parasite internalisation) thus allowing accumulation in the fully formed parasitophorous vacuole. A small proportion of RON12 appears to be present in the moving junction. RON12 does not appear to be essential but its deletion reduces parasite proliferation.
The motor behind the invasion process is an actinomyosin motor complex that is assembled below the parasite's plasma membrane.[128] This complex includes myosin, myosin tail domain interacting protein and glideosome associated proteins 45 and 50. It is anchored on the inner membrane complex which underlies the cell membrane. Myosin, myosin tail domain interacting protein and GAP45 first form a complex that then associates with GAP50. GAP45 is phosphorylated by calcium dependent protein kinase 1 on a number of serine residues. Removal of these residues does not appear to affect the assembly of this complex. This complex may have other function in addition to its role erythrocyte invasion.
The invasion process requires a coupling of the actin-myosin motor to the surface receptors. The myosin molecule involved belongs to the single-headed class XIV myosin. For the thromobospondin related anonymous protein on the sporozoites, aldolase which can bind actin forms this connection.[129] This connection requires tryptophan and negatively charged amino acids in the ligand's cytoplasmic tail. PfRH2b also binds aldolase with its cytoplasmic tail. This binning requires an aromatic amino acid (phenylalanine or tyrosine) rather than tryptophan again also in the context of negatively charged amino acids. PfLRH2a does not bind aldolase. A second protein glyceraldehyde-3-phosphate dehydrogenase can also bind actin. It is capable of biding the cytoplasmic tails of some of the PfRh and Duffy binding ligands in an aromatic amino acid dependent manner.
Trophozoite
After invading the erythrocyte, the parasite loses its specific invasion organelles (apical complex and surface coat) and de-differentiates into a round trophozoite located within a parasitophorous vacuole in the red blood cell cytoplasm. The young trophozoite (or "ring" stage, because of its morphology on stained blood films) grows substantially before undergoing schizogonic division.[130]
During asexual development the parasite increases in size to ~50% of the uninfected erythrocyte volume: the infected erythrocyte volume remains relatively constant.[131] Haemoglobin content gradually decreases but its concentration remains constant until the early trophozoite stage when it decreases by 25%. It then remains constant again until just prior to rupture. During early sexual development the gametocyte has a similar morphology to a trophozoite but subsequently undergoes a dramatic shape change.
The parasite's presence within the erythrocyte induces changes in the properties of the host cell. Relative membrane deformability is less than 10% of uninfected erythrocytes.[132] This change may contribute to the capillary occlusions that occurs in this disease. The deformability of the membrane is also dependent on the temperature and decreases with increased temperature. Deformability is reduced by a factor of 3-4 between 37 and 41 degrees Celsius. The fever that is commonly found in malaria may also contribute via this mechanism to capillary occlusion. The stiffness of the erythrocyte membrane increases as the parasite matures. The overall effect of these chances are to transform the erythrocyte from its normal biconcave shape into a parachute like structure. This change is apparent at high pressure rather than at low. Transition occurs at flow rates of ~65 µm per second. The mechanism of these changes are not known but changes in ATP consumption or alterations to the erythrocytes' spectrin framework may be important.
Within the red blood cell, the parasite metabolism depends greatly on the digestion of hemoglobin. A set of enzymes known as plasmepsins which are aspartic acid proteases are used to degrade hemoglobin. The parasite digests 70-80% of the erythrocyte's haemoglobin[133] but utilizes only ~15% in de novo protein synthesis.[134] Intraerythrocytic Plasmodium falciparum utilizes only a fraction of the amino acids derived from the digestion of host cell cytosol for the biosynthesis of its proteins. The excess amino acids are exported from the infected erythrocyte by new transport pathways created by the parasite.[135] The reason proposed for this apparently excessive digestion of haemoglobin is the colloid-osmotic hypothesis[136] which suggests that the digestion of haemoglobin increases the osmotic pressure within the infected erythrocyte leading to its premature rupture and subsequent death of the parasite. To avoid this fate much of the haemoglobin is digested and exported from the erythrocyte. This hypothesis has been experimentally confirmed.[137]
Infected erythrocytes are often sequestered in various human tissues or organs, such as the heart, liver and brain. This is caused by parasite derived cell surface proteins being present on the red blood cell membrane and it is these proteins that bind to receptors on human cells. Sequestration in the brain causes cerebral malaria, a very severe form of the disease, which increases the victim's likelihood of death.
The parasite can also alter the morphology of the red blood cell causing knobs on the erythrocyte membrane.
Erythrocyte invasion and growth leads to activation of several distinct anion channels and a non-selective Ca2+-permeable cation channel.[138] The non-selective cation channel's activation allows entry of Ca2+ and Na+. Absence of the channels is incompatible with pathogen survival. Although the mechanism of activation of these channels is not know it is presumed to be due to oxidation stree generated by the parasite because similar or identical channels are activated by oxidation of non-infected erythrocytes. Ca2+ entry stimulates an intraerythrocytic scramblase that facilitates bi-directional phospholipid migration across the bilayer. This results in an alternation of the cell membrane's phosphatidylserine asymmetry. Exposure of phosphatidylserine at the outer surface of the cell membrane is followed by binding to phosphatidylserine receptors on macrophages and the subsequent phagocytosis of the affected erythrocyte. It appears that the parasite because of its growth requirements is in a race to complete its life cycle before the infected erythrocyte is phagocytosed.
Schizont
At the schizont stage, the parasite replicates its DNA multiple times without cellular segmentation. These schizonts then undergo cellular segmentation and differentiation to form roughly 16-18 merozoite cells in the erythrocyte.[130] The rhoptries are formed mainly between second and fourth nuclear divisions; the micronemes between the end of the fourth nuclear division and merozoite separation from the residual body.[139] The dense granules are formed mainly after the micronemes.
The merozoites burst from the red blood cell, and proceed to infect other erythrocytes. The parasite is in the bloodstream for roughly 60 seconds before it has entered another erythrocyte.[58]
This infection cycle occurs in a highly synchronous fashion, with roughly all of the parasites throughout the blood in the same stage of development. This precise clocking mechanism has been shown to be dependent on the human host's own circadian rhythm.[140] Specifically, human body temperature changes as a result of the circadian rhythm, seem to play a role in the development of P. falciparum within the erythrocytic stage.
The synchronicity of the erythrocytic cycle is at least in part dependent on melatonin secretion by the host. A mechanism for this has been proposed.[141] Melatonin can activate phospholipase C which acts to generate inositol trisphosphate (IP3) which opens IP3 sensitive calcium channels in the endoplasmic reticulum. The released calcium in its turn controls the cycle through mechanisms that have yet to be understood.
Gametocyte differentiation
During the erythrocytic stage, some merozoites develop into male and female gametocytes - a process is called gametocytogenesis.[142] The specific factors and causes underlying this sexual differentiation are largely unknown. The base rate appears to be ~10% in laboratory strains but this is subject to many influences. One of the earliest suggestions in this area was made by Sinton who proposed that 'stress' might influence the rate of gametogensis.[143] There is some evidence from other workers to support this hypothesis.[144] Factors that have been reported to influence this rate include host erythrocyte age, hypoxia and exposure to schizonticidal drugs.
Gametocytes take roughly 8–10 days to reach full maturity and are metabolically active. The gametocytes remain within the erythrocytes until taken up by the mosquito host. The osmiophilic bodies, present in both male and female gametocytes but more abundant in the latter, are involved in the parasites' escape from the red blood cell during gametogenesis.[145][146]
There is considerable variation in the ratio of female to male gametocytes between strains.[147] Typically there are four female gametocytes to each male gametocyte. Mathematical modelling suggests that this may be due to differences in male fecundity.[148]
The protein Pfg377 has been shown to be localised to the osmophilic bodies of the gametocytes.[149]
The development of gametocytes is associated with down regulation of the erythrocyte membrane protein 1 (the product of the var genes) and loss of cytoadhesion.[150]
The CPW-WPC protein family, named after the unique WxC repeat domains, is highly conserved among Plasmodium species. It is transcribed in gametocytes and CPW-WPC proteins are expressed in zygote/ookinete-stage parasites.[151] These do not appear to be essential genes.
Sexual differentiation
Sexual parasite development is controlled by a DEAD box RNA helicase of the DDX6 family, termed DOZI.[30]
The Puf2 gene, a member of the Puf family of transcriptional regulators, has been shown to be involved in gamete formation.[152]
FACT (facilitates chromatin transcription) is a dimeric complex of two proteins - SPT16 and SSRP1 - which acts as a histone chaperone in the (dis)assembly of nucleosome (and chromatin) structure during transcription and DNA replication.[153] It is an essential gene in Plasmodium. Changing its promoter to one expressed only in the blood stages leads to changes in the male gametocytes. The mutant gametocytes have delayed DNA replication and gametocyte formation. Male gamete fertility is strongly reduced. Female gametocytes appear to be normal. When successful fertilization is achieved, the ookinetes generate oocysts that arrest early in development and fail to enter sporogony.
The proteins cell division cycle protein 20 and its homologue, CDC20 homologue 1 are central to the cell cycle activating the anaphase-promoting complex/cyclosome (APC/C) in mitosis and facilitating degradation of mitotic APC/C substrates.[154] A single homolog of this gene has been identified in Plasmodium berghei. It appears to be essential in male gametogensis but not for asexual reproduction. Blockage occurs at the nuclear spindle/kinetochore stage.
A gametocyte development 1 gene (Gdv1) which encodes a perinuclear protein has been identified.[155] Its mechanism of action is not known. Homologues of this gene have been found in Plasmodium vivax, Plasmodium knowlesi and Plasmodium gallinaceum.
Egress from the erythrocyte
This is an essential step in the life cycle. The calcium dependent protein kinase PfCDPK5 which is expressed in the merozoite is essential for this process.[156] Deletion mutations of this gene result in cell arrest in the late schizont stages. Merozoites released from these schizonts are capable of invasion.
The mechanics of release from the host membrane are partly known.[157] Before release a pore opens in the membrane. A distortion of the usual arrengement of the lipids and proteins of the host cell membrane occurs around the pore. This area of altered protein and lipid eventually ruptures.
Large holes appear in the cytoskeleton ~35 hours post invasion.[158] This occurs at the same time as the loss of cytoskeletal adaptor proteins that are part of the junctional complex, including α/β-adducin and tropomyosin. This is followed by the proteolysis of many cytoskeletal proteins during egress at ~48 hours post infection. This later proteolysis is mediated by the erythrocyte's own calpain-1.
Infected erythrocytes release microvesicles from their surface. They contain a number of parasite antigens associated with host cell membranes and proteins involved in parasite invasion which have potent immunomodulatory properties affecting macrophages and neutrophils.[159] Their uptake by infected eythrocytes stimulates the production of transmission stage parasites in a dose dependent manner. Their release increases during the asexual parasite cycle particularly prior to parasite egress. A protein - PTP2 - appears to be essential in this process but its role is not yet clear.[160]
A Gα(q) coupled signaling pathway that results in protein kinase C mediated loss of the host cytoskeletal protein adducin and weakening of the cellular cytoskeleton has also been implicated in the egress mechanism.[161] This weaking of the cytoskeletal induces catastrophic Ca2+ influx mediated by the mechanosensitive cation channel TRPC6. This in turn which activates host cell's calpain which proteolyzes the host cytoskeleton allowing parasite release.
Along with the release from the erythrocyte of the merozoites, the now functionless digestive vacuole is also released. These can active complement and are rapidly taken up by the polymorphs.[162] On ingestion the digestive vacuoles induce a vigorous respiratory burst which drives the cells into a state of functional exhaustion, blunting production of reactive oxygen species and microbicidal activity upon challenge with bacterial pathogens.
The serine repeat antigen (SERA) multigene family encode a series of proteins with a putative papain-like cysteine protease motif. One of these SERA5 (120 kiloDaltons) is produced at the late trophozoite/schizont stage. It is secreted together with other SERAs into the parasitophorous vacuole in an infected erythrocyte where it is cleaved into three fragments: an N-terminal domain (47 kDa), a central domain containing putative papain-like cysteine protease motifs (56 kDa) and a C-terminal domain (18 kDa). This N-terminal fragment is then cleaved in turn into two 25 kDa fragments. These fragments become covalently linked to the C-terminal 18 kDa fragment via disulfide bonding and attach to the merozoite surface. The central fragment is further cleaved to 50 kDa and 6 kDa fragments before being shed to the medium. These proteolytic cleavages are carried out by a subtilisin-like serine protease called PfSUB1 and the inhibition of this processing, likewise, results in blockade of merozoite release.[163] SERA6 may also be involved in schizont rupture and merozoite release from the erythrocyte. Both SERA5 and SERA6 are essential for blood stage parasite viability.[164] SERA6 is found in parasitophorous vacuole where it is activated by cleavage by the serine protease PfSUB1 just prior to egress. The release of PfSUB1 may be controlled by a calcium flux within the exomemes - storage vesicles within the parasite - of the merozoites.[165] The release may be under the control of a phospholipase C.
PfSUB1 - which is encoded by the gene PF3D7_0507500 - is released from the exomemes into the parasitophorous vacuole before the merozoite egresses from the erythrocyte. Its release is inhibited by a cyclic GMP dependent protein kinase (encoded by the gene PF3D7_1436600).[166] Substrates of PfSUB1 include MSP1, MSP6, MSP7, MSRP2, SERA5 and SERA6 and the rhoptry protein RAP1. PfSUB1 acts on MSP1 at three sites to produce 4 smaller proteins. It acts on SERA5 - located within the parasitophorous vacuole - a at one site to produce two smaller proteins. One of these is subsequently cleaved by a second protease. Inhibition of the cyclic GMP dependent protein kinase also inhibits microneme discharge.
A protein - gamete egress and sporozoite traversal - has been identified that appears to be involved in the egress of male and female gametes from the erythrocyte.[167] It is also involved in sporozoite migration.
A perforin-like protein (PPLP2) is involved in egress of male gametocytes from the erythrocytes.[168] This protein does not appear to be involved in the rupture of the parasitophorous vacuole. Instead of parasites with mutations in this gene produce gametocytes with only one, shared thicker flagellum rather than usual pattern of each male gametocyte forming 8 flagellated gametes.
Perforin-like protein 1 and perforin-like protein 2 are both transcribed in the blood stage.[169] Perforin-like protein 1 localizes to the red blood cell membrane and parasitophorous vacuolar membrane in mature schizonts following its Ca2+-dependent discharge from micronemes. Perforin-like protein 1 has Ca2+-dependent permeabilization and membranolytic activities.
The internal calcium levels rise for two hours pre egress.[170] This rise is dependent on internal stores of calcium probably from the endoplasmic reticulum. Inhibition of this rise prevents parasitophorous vacuole swelling and erythrocyte membrane poration.
Phosphorylation of the reticulocyte homologue protein 2b on residue on serine 3233 occurs prior to egress.[171] This is the sole phosphorylation site on this protein. This action is carried put by casein kinase 2 and is cyclic AMP independent, utilizes ATP as well as GTP as phosphate donors and is inhibited by heparin and tetrabromocinnamic acid. This phosphorlation most likely occus most likely occurs before the protein is translocated from the rhoptry neck to the plasma membrane.
Mosquito stage
P. falciparum is taken up by the female Anopheles mosquito as it takes its bloodmeal from an infected human. Within the midgut human complement remains active for ~1 hour. To protect itself the parasite binds host complement regulator factor H.[172] Specifically the gamete surface protein PfGAP50 binds to Factor H and uses the surface bound Factor H to inactivate the complement protein C3b.
How the parasite evades the mosquito's immune system is not understood. The protein Pfs47 appears to be involved in this process.[173]
Gametogenesis
Upon being taken up by the mosquito, the gametocytes leave the erythrocyte shell and differentiate into gametes. The female gamete maturation process entails slight morphological changes, as it becomes enlarged and spherical. On the other hand, the male gamete maturation involves significant morphological development. The male gamete's DNA divides three times to form eight nuclei. Concurrently, eight flagella are formed. Each flagellum pairs with a nucleus to form a microgamete, which separates from the parasite cell - a process known as exflagellation.
Gametogenesis has been shown to be caused by:[174]
- a sudden drop in temperature upon leaving the human host
- a rise in pH within the mosquito
- xanthurenic acid within the mosquito
An ATP-binding cassette (ABC) transporter encoded by the gene Pf14_0244 (PfABCG2) on chromosome 14 appears to have some role in the asexual stages, gametocyte stages and in the oocyst.[175]
The process of the formation of flagella is unusual in that they are formed with in the cytoplasm before export.[176] This pattern is also found in Chromera velia.
The movement of the flagellae has been studied.[177] The flagellae alternate their direction of beating on each stroke: a left to right beat is followed by a right to left beat.
Fertilization
Fertilization of the female gamete by the male gamete occurs rapidly after gametogenesis. The motility of the male gametocytes is powered by glycolysis.[178] The fertilization event produces a zygote. The zygote then develops into an ookinete. Of all the macrogametes present in the gut only ~2% develop into ookinetes.[179] The zygote and ookinete are the only diploid stages of P. falciparum.[citation needed]
A gene - PBANKA_113980 - essential for the production of viable progeny after meiosis has been identified.[180] This gene is the ortholog of the gene GEX1 in the plant Arabidopsis thaliana, the gene Cre06.g280600 in Chlamydomonas and the gene KAR5 in the yeast Saccharomyces cerevisiae. It appears that these genes are involved in nuclear fusion during fertilization.
Ookinete
The diploid ookinete is an invasive form of P. falciparum within the mosquito. It traverses the peritrophic membrane of the mosquito midgut and cross the midgut epithelium. Once through the epithelium, the ookinete enters the basal lamina, and forms an oocyst. Unlike the other invasive stages the ookinete lacks rhopteries. Only 1-2% of ookinites develop into oocyts.[179]
The processes of maturation and invasion of the mosquito gut are under investigation. Both chitinase and plasmepsin 4 (an aspartic acid protease) are known to be involved in the invasion process.[181]
During the ookinete stage, genetic recombination can occur. This takes place if the ookinete was formed from male and female gametes derived from different populations. This can occur if the human host contained multiple populations of the parasite, or if the mosquito fed from multiple infected individuals within a short time-frame.
A G-actin binding protein has been implicated in this process.[182] These proteins - probably acting as dimers - bind actin monomers just before sporogony.
Azithromycin has been shown to suppress apicoplast replication at the period of sporozoite production in oocysts.[183]
The proteins enolase and actin are present on the surface of ookinetes but their function there, if any, is unknown.[184]
The myosin gene (MyoA) class XIV is essential for ookinete motility in the mosquito. Mutants placed under a different promotor active in the blood stages failed to complete their life cycle.[185] Disruption serine repeat antigen 5 blocks parasite inhibits egress of sporozoites from an oocyst.[186]
Within the genome is encoded a protein phosphatase with a domain architecture known only otherwise to occur in Plantae: protein phosphatase containing kelch-like domains (PPKL).[187][188] The kelch motif normally occurs as a series of four to seven repeats forming a beta propeller tertiary structure and may occur at either terminus of the protein. In Plasmodium PPKL has five full and a one half kelch domains with 5 inserts unique to Alveolates. This protein is produced in schizonts and female gametocytes and is maternally inherited. Absence of this protein results in leads to the development of a malformed, immotile, non infectious ookinetes with extended apical protrusions. This protein is localised at the ookinete apical tip. Secretion of microneme contents is unaffected.
Development of the motile and invasive ookinete within the mosquito midgut is dependent upon two NIMA-related kinases, NEK2 and NEK4.[189][190]
Two potassium channel have been identified in the genome.[191] At least one of these is critical for the development of the ookinete in the mosquito.
The meiotic specific recombinase Dmc1 - a bacterial RecA homolog - appears to be essential for normal oocyst development in Plasmodium berghei.[192]
Sporogony
Over the period of a 1–3 weeks, the oocyst grows to a size of tens to hundreds of micrometres. During this time, multiple nuclear divisions occur. After oocyst maturation is complete, the oocyst divides to form multiple haploid sporozoites. The number of sporozoiter produced per oocyst varies but the mean is ~500-700 per oocyst.[179][193] Immature sporozoites break through the oocyst wall into the haemolymph. The sporozoites then migrate to the salivary glands and complete their differentiation. Once mature, the sporozoites can proceed to infect a human host during a subsequent mosquito bite.
Cell biology

Morphology
The nucleus, mitochondrion, apicoplast and the microtubules of Plasmodium sporozoites are linked to the parasite pellicle via long tethering proteins. The tethers originate from the inner membrane complex and are arranged in a periodic fashion following a 32 nanometer repeat. The tethers pass through a subpellicular structure that encompasses the entire parasite, probably as a network of membrane associated filaments.[194]
The volume of the nucleus of the asexual stages (~5 µm3) is approximately constant in size in the ring and trophozoite stages.[195] The nucleus is divided into several subcompartments including the nucleolus and the nuclear periphery which appear to be involved in transcription control.
The pellicle is a structure unique to the Apicomplexa and is made up of four components: the plasma membrane, the inner membrane complex, the subpellicular network and the subpellicular microtubules.[196] The subpellicular network consists of a two-dimensional network of intermediate filaments located on the cytoplasmic side of the inner membrane complex and acts as a membrane skeleton. The proteins - the inner membrane complex proteins (IMCs) - that compose this structure are functional homologs of the articulins, the membrane skeleton proteins of free-living protists.
The subpellicular network contains alveolins - a group of proteins that determine the shape of the parasite.[197] Another protein - G2 - which is structurally unrelated to alveolins appears to be involved in the organisation of the subpellicular microtubules.
Cell division
Cell division occurs through a process known as schizogony. This is a type of mitotic division in which multiple rounds of nuclear divisions occur before the cytoplasm segments.
DNA synthesis begins in the relatively small trophozoites but nuclear subdivision, which leads to the formation of multinucleate cells, occurs only during schizogony. Whether or not any gap phases exist between each round of DNA synthesis and mitosis is unknown. Eventually, a schizont composed of 8–32 nuclei undergoes segmentation, which culminates with the formation of individual merozoites that burst from the erythrocyte into the blood stream.
Apical complex
The rhopteries appear to have subcompartments allowing for differential secretion during the life cycle.[198] Two of these are known as the neck and the bulb.[199] A number of rhoptry neck proteins are conserved between apicomplexan species and are involved in host cell invasion. Bulb proteins in contrast are less well conserved between the apicomplexa and most likely evolved for a particular lifestyle. In the majority of species studied to date, rhoptry content is involved in formation and maintenance of the parasitophorous vacuole.
- Rhoptery proteins
The rhoptery neck proteins (RONs) along with the micronemal AMA1 protein are important in the penetration of the erythrocyte.[38] The invasion mechanism while common to all Apicomplexa is unique and involves a tight interaction between the host cell and the parasite surfaces called the moving junction. The moving junction, which is the anchoring structure for the invasion process, is formed by secretion of a macromolecular complex (proteins RON-2, -4, -5, -8), derived from the rhoptries, into the host cell membrane.
The mechanisms involved in this process are still being elucidated. The protein RON8 appears to be central to the binding of parasite to the erythrocyte surface.[200] Apical Sushi Protein and Rhoptry Neck protein 2 are released early following the formation of the tight junction between the merozoite and the erythrocyte. The rhoptry protein PFF0645c is released only after invasion is complete.
The P. falciparum apical sushi protein is the homolog of the P. vivax RON1 protein.[201]
The rhoptry protein 2 of Plasmodium vivax has been cloned.[202] The 1,369 amino acid protein is encoded PVX_099930 gene. The gene has nine introns and the protein contains a signal peptide at its N-terminus and 12 cysteines predominantly in its C-terminal half. It is localized in one of the apical organelles of the merozoite, the rhoptry, and the localization pattern is similar to its homolog in P. falciparum.
RON2 is inserted as an integral membrane protein in the host cell.[123]
The residues of the RON2 protein that bind to the AMA-1 protein have been identified.[203] It also appears that the formation of the junction and parasitophorous vacuole are molecularly distinct steps in the invasion process. Positive diversifying selection appears to have acted in the RON2 protein of Plasmodium vivax.[204]
In Plasmodium berghei RON4 is required for sporozoite invasion of hepatocytes.[38]
RhopH2 is transcribed and localised to the endoplasmic reticulum of the trophozoites by 28 hours post invasion.[205] This pathway is Brefeldin A-sensitive. By 32 hours the protein is distributed in the schizonts' cytoplasm but not in the parasitophorous vacuole.
A protein - Armadillo Repeats-Only - has been localised to the cytosolic face of the rhoptries.[206] A putative signal sequence in the first 20 amino acids has also been identified.
A number of heparin binding proteins are present in the rhopteries.[106] Heparin like molecules bound to the surface of the erythrocyte appear to be important in the adhesion process which also involves the merozoite surface protein 1.[63]
A protein S-acyl transferase with a DHHC-cysteine-rich domain is present in the rhoptry.[207] This enzyme may play a role in the apical positioning of the rhoptry organelles.
- Micronemal proteins
Apical membrane antigen-1 (AMA-1) - the product of the Pfama1 gene - is a surface exposed protein that plays a role in erythrocyte invasion. It is shed from the parasite surface predominantly via the action of the protease Sub2[208] Using gene deletion mutants AMA-1 has been shown to be a non essential protein.[209] While attached to the surface of the parasite AMA-1 can bind to RON2 - a protein that is inserted by the parasite into the erythrocyte membrane. This binding seems to be involved in the invasion process.
The receptor binding site of AMA-1 comprises the hydrophobic groove and a region that becomes exposed by displacement of the flexible domain II loop.[210]
Sub2 is released from the micronemes and can also act on the MSP1/6/7 complex and PTRAMP - another micronemal protein. Sub2 appears to be an essential gene.
- Proteins whose location is unclear
Several proteins are involved in the binding of the sporozoite to the various tissues it attaches to. TRAP, S6 and TLP have been implicated in these processes.[211]
Maurer's clefts
In 1902 the German physician Georg Maurer discovered an unusual staining pattern in the cytoplasm of erythrocytes infected with P. falciparum. These structures were subsequently named Maurer's clefts. These consist of a convoluted set of membranes that lie within the erythrocyte's cytoplasm and appear to be involved in secrection from the erythrocyte.[212] They are known to have proteins of parasite origin within them including the Maurer's cleft two transmembrane proteins (PfMC-2TM)[213] The clefts appear to originate from vacuoles budding off them the parasitophorous vacuole membrane which then diffuse within the erythrocyte cytoplasm before taking up residence at the cell periphery.[214]
Another protein associated with these structure is skeleton-binding protein 1 (SBP1). This protein is involved in transport of the var gene protein, PfEMP1 (erythrocyte membrane protein 1) to the erythrocyte surface.[215]
Other proteins associated with these structures include membrane associated histidine rich protein 1 and ring exported protein 1 and 2.[216][217]
Mutations in the ring exported protein 1 (Rex 1), a protein normally found in Maurer's clefts, reduces transport of the var gene products to the erythrocyte surface.[218]
The erythrocyte protein ankyrin is found in these structures.[219]
The parasite generates a host derived actin cytoskeleton within the cytoplasm of the erythrocytes that connects the Maurer's clefts with the host cell membrane and to which transport vesicles are attached.[220] Hemoglobin oxidation products which are enriched in hemoglobin S and C containing erythrocytes inhibit actin polymerization. This may account for their protective role in malaria.
The protein trophozoite exported protein 1 (PFF0165c) is located within the clefts.[221] The protein's N-terminal region is intrinsically unstructured but it also has a coiled coil domain. It appears to lack export motifs such as PEXEL, signal sequence/anchor or a transmembrane domain. Transport of this protein to the clefts is sensitive to inhibition by Brefeldin A. This is normally associated with proteins that are have co-translation translocation into the endoplasmic reticulum or posttranslational insertion into the endoplasmic reticulum followed by vesicular transport from the endoplasmic reticulum via Golgi apperatus to the cell surface.
Mutations in REX1 and Pf332 proteins result in distortion of Maurer's clefts morphology suggesting that they play a role in its structure.[222][223]
The actin network exerts skeletal functions by anchoring the Maurer's clefts within the erythrocyte cytoplasm and restrains the Brownian motion of this organelle.[224] Haemoglobin S and C appear to interfere with this organisation.
The protein Surfin 4.1 is found in the clefts.[225]
The membrane associated histidine rich protein 2 is attached to Maurer's clefts.[226]
There are three multigene families organized into 9 highly conserved clusters with the Pfmc-2tm genes in the subtelomeric regions of the chromosomes.[227] These genes are expressed at early trophozoite stages. Like the PfMC-2TM proteins, the PfEPF1, 3 and 4 proteins encoded by these families are exported to the Maurer's clefts, as peripheral or integral proteins of the Maurer's cleft membrane and largely exposed to the red cell cytosolic face of this membrane.
Within the PfMC-2TM proteins there is a conserved domain MC-TYR.[228]
Dense granules
These are small vacuoles that can be seen on electron microscopy. They bind osmium and appear dark on the images. Little is known about these organelles but they are thought to play a role in the maintenance of the parasitophorous vacuole. One protein - p377 - has been localised to these organelles. Disruption of this protein reduces osmiophilic body formation, and leads to a marked decrease in female fitness and impaired infectivity to mosquitoes.[229]
A protein - the ring membrane antigen (RIMA) - has been localised to the dense granules.[230] The protein's molecular weight is 14 kiloDaltons and it is synthesized late in schizogony. At the late schizont stage this protein is distributed diffusely throughout the intracellular schizont. During the segmenter stage it is then localized to the dense granules.
Parasitophorous vacuole
Within a red blood cell, P. falciparum resides inside the parasitophorous vacuole. This is formed during erythrocyte invasion.
The proteins originating in the parasite pass through the membrane of the parasitophorous vacuole and are transported to the cytoplasm or membrane of the erythrocyte.[3] Although this transport mechanism is largely unknown some details have been elucidated.[231] Ingestion of the erythrocyte cytoplasm begins in mid-ring-stage parasites. Host cytoplasm is internalised via cytostome-derived invaginations and then concentrated into several acidified peripheral structures. Haemoglobin digestion and haemozoin formation occur within these vesicles. The ring-stage parasites can adopt a deeply invaginated cup shape, but they do not take up haemoglobin via macropinocytosis. As the parasite matures the haemozoin containing compartments coalesce to form a single acidic digestive vacuole (pH 4.5 - 5.5) that is then fed by haemoglobin containing vesicles. Some haemoglobin degradation also occurs in compartments outside the digestive vacuole.
The enzyme phosphatidyl-inositol-3-kinase (PI3K) has been implicated in this process.[232] PI3K is located in vesicular compartments near the membrane and in the digestive vacuole and is involved in endocytosis from the host and trafficking of hemoglobin in the parasite. Its inhibition with wortmannin or LY294002 results in entrapment of hemoglobin in vesicles within the parasite cytoplasm preventing its transport to the digestive vacuole.
The pH of the digestive vacuole is maintained by a V-type H(+)-ATPase.
A signal sequence at the N terminal of proteins targeted to the arasitophorous vacuole has been identified.[233] The signal appears to reside in the 55 amino acids of the N terminal of the protein. There may be a retention signal at the C terminal.
The micronemal protease ROM1 appears to be essential for proper parasitophorous vacuole modification to allow parasite development.[234] This protease is able to cleave the proteins AMA1 and MAEBL.
Uric acid precipitates are present in the cytoplasm of the parasitophorous vacuole.[235] These are released when the merozoites are released. Uric acid is highly inflammatory and can cause maturation of dendritic cells.
The protein PFA0210c is a member of the START domain family which is involved in the transport of phospholipids, ceramide or fatty acids between membranes.[236] It associates with membranes in infected erythrocytes at mature stages of intracellular parasite growth. It is present in the parasitophorous vacuole during growth and is later recruited to organelles in the parasite.
Apicoplast
Plasmodium falciparum, and most other members of the phylum Apicomplexa, contain an organelle termed an apicoplast.[3] It was first discovered in 1996.[237] The apicoplast is an essential plastid, homologous to a chloroplast, although the apicoplast itself lacks any photosynthetic function. Evolutionarily it is thought to have been derived through secondary endosymbiosis. As humans do not harbor apicoplasts, this organelle and its constituents are seen as a possible target for antimalarial drugs.
The apicoplast varies in size during the life cycle from ~0.5 µm × 0.15 µm in the merozoite to 1.6 µm × 0.35 µm in the trophozoites.[53] Only one copy is present until it replicates in late schizonts. The apicoplast always adheres to the mitochondrion, along its whole length in merozoites and early rings but only at one end in later stages. Regions of the apicoplast are also closely related to the pigment vacuole, nuclear membrane and endoplasmic reticulum. In merozoites the plastid is anchored to a band of 2-3 subpellicular microtubules.
The apicoplast has four membranes is normally located between the nucleus and the rhoptries.[238] Its matrix contains ribosome sized particles and membranous whorls. The gap between the second and third membranes is frequently larger than between the other membranes. The interior matrix of the apicoplast contains ribosome-like granules and a network of fine branched filaments.
It contains a 35-kb genome, which encodes for 30 proteins. The genome of this organelle has now been sequenced for several species.[239] It appears to be conserved and to encode ~30 genes in all species examined.
The plastid genome replicates at the late trophozoite stage of the parasite intraerythocytic cycle. It proceeds predominantly via a D-loop/bi-directional ori mechanism with replication ori localized within the inverted repeat region. The process of replication involves a nuclear-encoded DnaJ homolog that binds to the ori site.[240]
The DNA polymerase involved in the replication of its genome is Pfprex (Klenow-like polymerase). This enzyme has been cloned, expressed and purified.[241] The enzymes is relatively error prone and shows a bias toward T->C mutations.
Other nuclear encoded proteins are transported into the apicoplast. Transport into the apicoplast are not well understood. These proteins has a signal in the N terminal but unlike many other organisms this appears to be a disordered chain rather than a conserved sequence.[242] It was thought that a specific signal peptide was responsible for this targeting [3] and it was estimated that 551, or roughly 10%, of the predicted nuclear-encoded proteins are targeted to the apicoplast. This hypothesis now appears to be incorrect. It appears that a relative enrichment within the protein of positively charged amino acid residues (Arginine, Histidine, Lysine) particularly at the N terminal of the protein may be sufficient to target the protein to the apicomplast.[243]
The biosynthesis of this organelle is not well understood. Phosphatidylinositol 3-monophosphate has been shown to be involved in its biosynthesis in the apicomplexian Toxoplasma gondii.[244] It seems likely that this enzyme is involved in the formation of this organelle in the Plasmodium species also.
This organelle appears to be essential in the liver stages.[245]
The functions of this organelle remains to be fully determined but it appears to be involved in the metabolism of fatty acids, isoprenoids and heme.[3] There are two pathways for protein lipoylation in Plasmodium - one in the mitochondrion and the other in the apicoplast. The apicoplast pathway is not found in the vertebrate host and relies on de novo lipoic acid synthesis.[246]
The role of the apicoplast in the blood stages has been clarified.[247] Inhibition of isoprenoid precursor biosynthesis with the antibiotic fosmidomycin (an inhibitor of the enzyme DOXP reductoisomerase) causes delayed death in this parasite. This effect can be overcome with the addition of isopentenyl pyrophosphate (IPP) to the culture medium. Continued culture in the presence of this agent leads to the loss of the apicoplast genome and these mutants fail to process or localize organelle proteins. These auxotrophs can be grown indefinitely in asexual blood stage culture but are entirely dependent on exogenous IPP for survival.
Iron-sulphur prosthetic groups are assembled in this organelle.[248] One component (SufB) is encoded in the apicoplast genome and a second (SufC) is encoded in the nucleus. SufB also exhibits ATPase activity. Other pathways that have been linked to this organelle include biosynthesis of isoprenoid precursors, fatty acids, heme and lipoic acid.[249]
Iron-sulfur (Fe-S) cluster cluster synthesis pathways are found in the apicoplast.[250] In the Suf pathway, SufS - a cysteine desulfurase - and its partner SufE are found exclusively in the apicoplast. In the Isc pathway IscS and its effector Isd11 were solely mitochondrial. Interruption of the Suf pathway results in a phenotpe that is dependent on external isopentenyl pyrophosphate. This phenotype is also associated with the loss of the apicoplast organelle and its organellar genome suggesting a role for this pathway in the maintenance of the apicoplast itself.
The [Fe-S] cluster protein NFUapi - a protein with a nitrogen fixation factor U (NifU)-like domain - is localised to the apicomplast.[251] This protein may have a role in merosome formation and appears to be non essential for the blood stages.
A gene Plasmodium-specific Apicoplast protein for Liver Merozoite formation (PALM) has been shown to be important for merozoite formation.[252] Knock out mutants are unable to release merozoites into the blood from the liver stages. Mutants lacking this gene appear to be able to elicit at least temporary immunity.
Falcilysin a zinc metalloprotease is found in the apicoplast.[253] It is a member of the M16 protease group and has maximal activity at neutral pH. It appears to be an essential gene. Its function in this organelle is not quite clear but it appears to be involved in the degradation of transit peptides.
The enzyme thioredoxin peroxidase is found in the apicoplast, the mitochondrion and the cytosol.[254]
Autophagy is membrane-mediated degradation process that involves a series of proteins known as Atg proteins. Atg8 is expressed during development and localises to the apicoplast.[255]
Two C3 sugar phosphate transporter are present in the membrane of the apicoplast of Plasmodium berghei - triose phosphate transporter and phosphoenolpyruvate transporter.[256] Knock out mutants of the triose phosphate transporter fail to survive. Phosphoenolpyruvate transporter knock out mutants survive in the blood stages but suffer defects during the liver stages development.
A pyruvate dehydrogenase complex with four subunits (E1alpha, E1beta, E2, E3) is present in the apicoplast.[257] Unlike plants there is only a single copy of these genes in the genome. pyruvate dehydrogenase deficient parasites have no apparent blood stage growth defect, they are unable to progress beyond the oocyst phase of the parasite's mosquito stage.[258]
An ATP dependent caseinolytic protease (ClpP) is present in the apicomplast. Its function is currently unknown.[259]
The lipid composition of the apicoplast has been analysed.[260] some apicoplast lipids are generated de novo by the organelle itself. Phosphatidylinositol and other phospholipids in the organelle are enriched in saturated fatty acids. Lipids atypical for plastids - sphingomyelins, ceramides and cholesterol - are present. Galactoglycerolipids, dominant in plant and algal plastids, are not present suggesting that these glycolipids are a hallmark of photosynthetic plastids and were lost when these organisms assumed a parasitic lifestyle.
The endoplasmic reticulum associated degradation (ERAD) machinery is a quality control mechanism that retro-translocates misfolded secretory proteins across the endoplasmic reticulum membrane.[261] Several components of this system including the membrane protein Der1, the AAA ATPase Cdc48 and its cofactor Ufd1 appear to be present in the apicoplast and are involved in transmembrane transport.
ClpB is a molecular chaperone and a member of the AAA+ superfamily of ATPases. There are 2 isoforms of ClpB (PfClpB1 and PfClpB2) found in the apicoplast.[262] PfClpB1 contains all characteristic AAA+sequence motifs but there is a 52-residue long non-conserved insert middle domain. ATP induces self association of PfClpB1 into hexamers like in most AAA+ ATPases. It catalyzes the hydrolysis of ATP and its ATPase activity is activated in the presence of casein and polylysine.
A glutamyl-tRNA synthetase (GluRS) is present in the apicoplast.[263] It is it is non-discriminating glutamylating both apicoplast tRNA-Glu and tRNA-Gln. It appears to be an essential enzyme.
An elongation factor G which is involved in translation is present in the apicoplast.[264] It has GTPase activity that can be inhibited by fusidic acid.
Mitochondrion
Plasmodium lacks mitochondrial pyruvate dehydrogenase[265][266] and the hydrogen ion translocating NADH dehydrogenase (Complex I, NDH1). The mitochondrion contains a minimal DNA genome (~6 kilobases) and carries out oxidative phosphorylation in the insect vector stages by using 2-oxoglutarate as an alternative means of entry into the tricarboxylic acid cycle and a single-subunit flavoprotein as an alternative NADH dehydrogenase (NDH2). In the blood stages mitochondrial enzymes are down regulated and parasite energy metabolism relies mainly on glycolysis. The enzyme malate quinone oxidoreductase was acquired from an epsilon proteobacteria via lateral gene transfer. This transfer occurred in an ancestor of the Apicomplexa.
The ATP synthase is localised to the mitochondrion, is assembled as a large dimeric complex and appears to be essential for in the blood stages of the life cycle.[267] Its function in these stages is not yet clear.
The sulfhydryl:cytochrome c oxidoreductase Erv1/ALR/GFER/HSS (Essential for Respiration and Vegatative growth/Augmenter of Liver Regeneration/Growth Factor Erv1-like/Hepatic regenerative Stimulation Substance/hepatopoietin) is an essential sulfhydryl oxidase for required oxidative protein import into the mitochondrial intermembrane space. It is one of several enzymes involved in electron transferase activity. It is encoded by all eukaryotes and cytoplasmic DNA viruses sequenced to date. The enzyme from P. falciparum differs significantly from that found in yeast and humans with altered cysteine motifs and intermolecular disulfide bonds.[268] Despite successful cloning and expression in yeast, the parasite enzyme fails to function in yeast. A second related enzyme - Mia40 - does not appear to be present in P. falciparum.
Deletion of the gene in the rodent parasite Plasmodium berghei for the flavoprotein subunit of succinate dehydrogenase - part of the complex II - showed impairment of ookinete function and oocyst formation.[269]
The gene for the flavoprotein subunit of succinate dehydrogenase can be disrupted in the parasite.[270] Its disruption causes growth retardation of the intraerythrocytic forms. It appears that complex II functions as a quinol-fumarate reductase to form succinate from fumarate in the intraerythrocytic parasite.
The dicarboxylate-tricarboxylate carrier homolog has been cloned from P. falciparum.[271] This protein may mediate the oxoglutarate-malate exchange across the inner mitochondrial membrane required for the branched pathway of tricarboxylic acid metabolism.
The ClpQ protease and ClpY ATPase have been cloned.[272] ClpQY function disruption caused hindrance in the parasite growth and maturation of asexual stages of parasites. Features of apoptosis like cell death are also found.
The mitochondrial pathway of protein lipoylation relies on scavenging from the host and can be inhibited with the lipoic acid analog 8-bromo-octanoic acid. Use of this agent inhibits growth and significantly reduces merosome formation. Schizogony is the phase most affected by this inhibition.[246]
Atovaquone, a 2-hydroxynaphthoquinone, is a competitive inhibitor of the quinol oxidation site of the mitochondrial cytochrome bc1 complex and is used as an antimalaria agent.[273] Inhibition of this enzyme leads to the collapse of the mitochondrial membrane potential and disruption of pyrimidine biosynthesis. These effects are lethal to the parasite.
The mitochondrial RNA polymerase appears to be an essential gene for the erythrocytic stages.[274]
Over half the genome of the mitochondrion encodes the genes for three classic mitochondrial proteins: cytochrome oxidase subunits I and III and apocytochrome b.[275] The remainder encodes 34 RNA genes of which 27 have been assigned to ribosomal RNA (12 to the small subunit and 15 to the large subunit). These genes are fragmented and are encoded on both strands.
The mitochondrial thioredoxin peroxidase-2 does not appear to be essential.[276]
The ATP dependent ClpQY system is a prokaryotic proteasome like multisubunit machinery localized in the mitochondrion. It has two components - a ClpQ threonine protease and ClpY ATPase. The ClpQ threonine protease appears to be an essential gene.[277]
The bc 1 complex of the mitochondrial respiratory chain is essential for the parasite's life cycle. The drug atovaquone inhibits its activity. Atovaquone binds to the Q0 site on this complex.[278]
An AAA+/FtsH protease homolog (PfFtsH1) that exhibits ATP- and Zn2+-dependent protease activity is present in the inner mitochondrial membrane.[279] It seems likely that this is a regulatory protein for organelle biogenesis.
An elongation factor G involved in protein translation is present in the mitochondrion.[264] It has GTPase activity that can be inhibited by fusidic acid.
Digestive vacuole
During growth of the parasite and as part of its digestion of the erythrocyte's haemoglobin, fusion of digestive vesicles occurs and gives rise to a large digestive vacuole.[280] This vacuole the interior of which is maintained at a low pH (pH 4.5 - 5.5), processes 60-80% of the ingested hemoglobin and provides a pool of amino acids that is crucial for parasite growth and development. The membrane contains ion pumps and transporters that maintain its low pH. During haemoglobin digestion the heme is released from hemoglobin. Haem is toxic to the parasite and is detoxified by biocrystallization to hemozoin within the vacuole. Quinoline drugs, including chloroquine, act by binding to heme and thus prevent its sequestration into hemozoin.
It has been shown that micromolar concentrations of chloroquine partially permeabilized the parasite's digestive vacuole membrane and that this event appears to precede mitochondrial dysfunction.[281]
Quinine has been shown localise to a non acidic compartment within the digestive vacuole.[282] It may colocate with haemozoin. It's localisation within the parasite is not altered by the presence or absence of a functional multidrug resistance gene.
The digestive vacuole is able to activate both the alternative complement and the intrinsic clotting pathway.[283] The digestive vacuole membrane has the capacity to assemble prothrombinase, a key enzyme of the intrinsic clotting pathway.[283] The capacity of this membrane to activate both complement and coagulation can be suppressed by low molecular weight dextran sulfate. Phagocytosis of these membranes drives the polymorphonucleocytes into a state of functional exhaustion.
Two multi-spanning digestive vacuole membrane proteins are known: the multidrug resistance protein 1 and Chloroquine Resistance Transporter (CRT).[284] The CRT protein moves from the endoplasmic reticulum to the Golgi apparatus before becoming associated with the digestive vacuole. The digestive vacuole forms in the ring stages of the parasites life cycle. Chloroquine sensitivity is not influenced by the absence of CRT from the digestive vacuole bringing into question its relationship (if any) to chloroquine resistance.[284] Mutations in the CRT gene have been associated with sensitivity and resistance to quinolines.[285] In particular a lysine to isoleucine at codon 76 (Adenosine -> Thymine at base 227) mutation and a valine to phenylalanine (Guanine -> Thymine at base 1108) mutation have been associated with changes in drug sensitivity. The mutant CRT protein increases the transport of glutathione into the digestive vacuole. The role - if any - that this has on chloroquine resistance is presently unknown.
The membrane of the digestive vacuole is four nanometers in thickness with patches that may be up to 12 nanometers in thickness.[286]
The digestion of haemoglobin produces large quantities of ferriprotoporphyrin IX which it unable to digest and is potentially toxic to the parasite. To avoid the toxicity the ferriprotoporphyrin is converted to haemozoin. Chloroquine inhibits this process. The mechanisms behind this process are still unclear. In vitro conversion of ferriprotoporphyrin to haemazoin is enhanced at a temperature of 41C when compared to its conversion at 37C.[287] It is possible that the rise in temperature that occurs in malaria may be part of a strategy to enhance this reaction at the later stages of growth when the ferriprotoporphyrin concentration is likely to be high.
Inhibition of the falcipains involved in the digestion of haemoglobin results in enlargement of the digestive vacuole.[288]
Nucleus
Unlike most other eukaryotes the chromosomes never become condensed even during mitosis and remain difficult to visualise by llight microscopy.
Studies with antibodies to the nuclear pore protein subunit PfNup116 show that the nuclear pores are highly polarized during the ring and schizont stages whereas in trophozoite stage the nuclear pores redistributed over the entire nuclear surface.[289]
PfSec13 is a nucleoporin.[290] It appears to be a fusion between Sec13 and Nup145C and it associates with the nuclear pore complexes and microtubules. It appears to be an essential gene.
Nucleolus
A hat-like structure polarized towards one side of the nucleus that stains with nucleolar markers has been described[291] It seems likely that this unusual structure is the nucleolus.
The histone deacetylase Sir2a is found in the nucleolus in addition to the telomeres.[292] It functions there to control the transcription of the ribosomal RNA levels.
The ribosomal RNA genes are found localised to this organelle.[293]
Endoplasmic reticulum
This forms a set of reticular structures adjacent to the nuclear regions during the trophozoite and schizont stages.[294] In the late schizont stage it forms globular structures surrounding each budding merozoite.
A Ca2+ ATPase 6 has been associated with resistance to artemisinin resistance.[295] The gene has a single copy in the genome.[296] Reistance has been associated with four mutations: codon 263 Lysine->Glutamic acid, codon 431 Glutamic acid->Lysine, codon 623 Alanine->Glutamic acid and codon 769 Serine->Asparagine.[297] This gene is a SERCA pump and appears to be essential in the asexual stages.[298]
Many proteins in the genome carry a host targeting signal.[299] This signal sequence is recognised by phosphatidylinositol-3-phosphate in the endoplasmic reticulum.
Ribosomes
Unlike other eukaryotes studied to date Plasmodium species have two or three distinct SSU rRNA (18S rRNA) molecules encoded within the genome.[300] These have been divided into types A, S and O. Type A is expressed in the asexual stages; type S in the sexual and type O only in the oocyte. Type O is only known to occur in Plasmodium vivax at present. The reason for this gene duplication is not known but presumably reflects an adaption to the different environments the parasite lives within.
The Asian simian Plasmodium species - Plasmodium coatneyi, Plasmodium cynomolgi, Plasmodium fragile, Plasmodium inui, Plasmodium fieldi, Plasmodium hylobati and Plasmodium simiovale - have a single single S-type-like gene and several A-type-like genes. Phylogenetic analyses has shown that gene duplication events giving rise to A- and S-type-like sequences took place independently at least three times in the Plasmodium evolution.
The phosphoprotein P0 occurs as a complex with two other small acidic ribosomal proteins (P1 and P2).[301] A pentameric complex [(P1–P2) P0 (P1–P2)] form the stalk of the large ribosomal subunit, which seems to play a role in the GTPase elongation centre of the ribosome.
The P2 protein is exported to the infected erythrocyte surface at 30 hrs post merozoite invasion, concomitant with extensive oligomerization. It is largely largely composed of alpha helical and random coil domains.[301]
Acidocalcisomes
Acidocalcisomes are acidic calcium stores and are present in many organisms including bacteria, Plasmodium and humans.[302] The organelles possess an acidic matrix that contains several cations bound to phosphates. These are mainly present in the form of short and long polyphosphate chains. The matrix is acidified through the action of proton pumps such as a vacuolar proton ATPase and a vacuolar proton pyrophosphatase. Calcium uptake occurs through a Ca2+/H+ countertransporting ATPase located in the membrane of the organelle.
Motility
Thrombospondin Related Anonymous Protein (TRAP) is a type I transmembrane proteins which has several extracellular adhesive domains and a cytoplasmic domain that recruits the glycolytic enzyme aldolase. Normally only small amount of TRAP found on the sporozoite surface. TRAP is involved in cell motility.[303]
Its tandem von Willebrand factor A and thrombospondin type I repeat domains connect through the proline rich stalk, transmembrane and cytoplasmic domains to the parasite's actin dependent motility apparatus.[304] Binding is dependent on the presence of a metal ion. The protein is capable of considerable conformational changes. There is also a potential heparan sulphate binding site in the von Willebrand factor A domain.[305]
The cytoplasmic domain binds to F-actin which connects to myosin A. Within the transmembrane domain it has a canonical rhomboid cleavage site (Ala-Gly-Gly-Ile-Ile-Gly-Gly). Rhomboid proteases are a family of serine proteases that require helical instability in the transmembrane domain and have specific residue requirements in their P1, P4 and P2′ positions. These proteases are responsible for intramembraneous cleavage.
TRAP binds to receptors on the host and is translocated posteriorly by the actomyosin motor. It is then normally cleaved by a calcium independent serine protease. Removal of the cytoplasmic domain abolish the motility of the parasite. Mutations in the rhomboid cleavage site are defective in TRAP shedding and display slow, staccato motility and reduced infectivity.[306] The reduction in infectivity is particularly marked if the sporozoites are inoculated intradermally rather than intravascularly. Prevention of cleavage of the TRAP protein entirely renders the sporozoites uninfectious and immobile. The rhomboid protease normally involved in TRAP cleavage appears to be the ROM4 protease. This protease is found across the entire sporozoite surface suggesting it has functions in addition to TRAP cleavage.
The circumsporozoite- and thrombospondin-related adhesive protein (CTRP) is a modular multidomain protein containing six tandem von Willebrand factor A like domains and seven tandem thrombospondin type I repeat-like domains.[307] The A domains of CTRP are critical for ookinete gliding motility and oocyst formation. The thrombospondin domains are fully redundant.
The cell-traversal protein for ookinetes and sporozoites (CelTOS) is a protein involved in the invasion of both vertebrate and insect host cells.[308]
The C-terminal tail of myosin A (MyoA) and its light chain, myosin A tail domain interacting protein (MTIP) are essential parts of the gliding motility apperatus.[309]
Dynein light chain 8 is present in P. falciparum as a homodimer.[310] The dimer is formed by the interaction of the β0 chains on one molecule with the β2 chains of the second.
A kinesin with the ability to depolymerise microtubules has been cloned.[311]
Autophagy
A number of proteins involved in autophagy are known to be present in the genome. These include Atg8 and Atg3.[312] The functions of these proteins in the parasite are still being elucidated.
Autophagosomes fuse with the endosomes before being routed to the digestive vacuole.[313] The digestive vacuole is probably used for this purpose because the parasite lacks lysosomes.
Crystaloids
These are transient structures whose presence is restricted to the mosquito specific ookinete and young oocyst stages of the parasite.[314] They are cytoplasmic aggregations of closely packed spherical particles 25–35 nanometers in diameter and disappear after ookinete-to-oocyst transition. Although first described in the 1962 their function is unknown.[315] They contain a scavenger receptor like protein which is formed in the macrogametocytes.[316] They also contain a number of LCCL proteins.[317]
Effect of radiation
Gamma rays may be used to produce attenuated parasites. Despite this the effects on the parasite have rarely been studied.[318] Gamma irradation acts in a dose dependent fashion: morphologically it induces defective mitosis, sparse cytoplasm, fewer ribosomes, disorganized and clumped organelles and large vacuoles. The transcription of a number of genes is altered.
Molecular biology and biochemistry
Redundancy of binding/invasion proteins
Proteins involved in this process are subject to three levels of host cell selection:
- selection between host species (species specific tropism)
- selection among individuals within a host species (erythrocyte receptor diversity)
- selection of subpopulations within an individual (age dependent invasion)
These proteins are necessarily surface exposed and are also subject to selection by the host's immune system
Because of the multiple levels of selection it is to be expected that a number of proteins are involved in this process. This also in part explains the apparent redundancy of these proteins.
Surface exposed proteins
- Merozoite surface protein (MSP) family
The cleavage of MSP 1 appears to involve a purinergic signalling pathway.[319]
The MSP1 protein binds the pro inflammatory protein S100P.[320] This binding appears to prevent the usual NFκB activation in monocytes and chemotaxis in neutrophils. S100P appears to be able to bind to at least 2 alleles of MSP1 which are separated by at least 27 million years of evolution suggesting that this inhibition mechanism may also be of considerable age.
Merozoite surface protein 2 is one of the most abundant proteins on the surface of merozoites, is intrinsically unstructured and forms amyloid-like fibrils in solution.
Merozoite surface protein 7 appears to enhance the virulence of the parasite at least in the rodent.[321]
A protein PfMSPDBL1 (encoded by PF10_0348 gene) that is a member of the MSP3 family and has both Duffy binding-like (DBL) domain and secreted polymorphic antigen associated with merozoites (SPAM) domain appears to be critical for erythrocyte invasion.[322] The merozoite surface proteins DBL1 and -2 (PfMSPDBL1 and PfMSPDBL2) (PF10_0348 and PF10_0355) are extrinsically associated with the merozoite. MSPDBL2 appears to have a role in resistance to halofantrine, mefloquine and lumefantrine.[323]
- Circumsporozoite protein (CSP)
The circumsporozoite protein (CSP) forms a dense coat on the sporozoite's surface.[324] It consists of approximately 400 amino acids organized into three domains: an N-terminal domain containing a conserved pentapeptide (region I), a highly repetitive species specific central domain and a C-terminal domain containing a second conserved sequence (region II). It is involved in invasion of the mosquito's salivary glands and the binding sporozoites to liver cells.
The circumsporozoite protein has been shown to be an inhibitor of the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB).[325] Its nuclear localization signal alone is sufficient to block NF-κB activation.
CSP binds to salivary glands and is involved in its invasionIn by the sporozoites. An Anopheles salivary gland protein - CSP-binding protein - to which CSP binds during this process has been identified.[326]
- Early transcribed membrane protein (ETRAMP) family
The ETRAMP family is characterized by a predicted signal peptide, a short lysine rich stretch, an internal transmembrane domain and a highly charged C-terminal region of variable length. The highly charged terminal region appears to be involved in protein-protein interactions.[327] They are usually expressed in a stage-specific manner. In the blood stages they localize to the parasitophorous vacuole membrane and to vesicle like structures exported to the host erythrocyte cytosol. The gene ETRAMP 10.3 has been shown to be expressed in the liver, sporozoites and blood stages.[328] Within the liver and blood stages it is localized to the parasitophorous vacuole membrane. It is also exported to the erythrocyte during the blood stages. It appears to be an essential gene in the blood stages.[329] In Plasmodium berghei two members of the ETRAMP family (uis3 and uis4) localize to secretory organelles of sporozoites and to the parasitophorous membrane vacuole of the liver stages.[330] Another member of this family - SEP2 - is expressed in the gametocytes, in the mosquito and in the liver stages.[330] In the liver stage SEP2 is routed to the parasitophorous vacuole membrane. In the ookinete and sporozoite stages, it instead localizes to the parasite surface. It is also released during gliding motility of salivary gland sporozoites.
- 6-cys domain proteins
A group of proteins known as the 6-cys domain proteins - so called because they contain modules with six characteristic cysteines forming three intra-molecular disulphide bonds between C1 and C2, C3 and C6, and C4 and C5 - are surface exposed proteins.[331] The first P12 - named after the clone it was isolated from - was described in 1990.[332] There are at least nine members of the 6-cys family. Most family members contain two 6-cys modules, but up to seven modules can be found in a single protein, in addition to incomplete modules containing fewer cysteine residues. About half of the 6-cys family members characterised to date possess glycosylphosphatidylinositol (GPI) moieties that anchor them to the outer leaflet of the plasma membrane, while those that lack GPI-anchors presumably remain associated with the parasite surface via interactions with other membrane proteins. Of this family P12, P38 and P41 are blood stage antigens. P230 and P48/45 - another two members of this family - are expressed on the surface of gametes.
P12 has only two s48/45 domains while other members have up to fourteen.[125] Pf12 is highly conserved and under purifying selection. It forms a heterodimeric complex with Pf41.
- LCCL/lectin adhesive-like protein family
There are a family of LCCL/lectin adhesive-like protein (LAP) proteins encoded in the genome.[333] The six members are expressed in gametocytes and form a multi-protein complex. There are normally six of these proteins in the genome. They are essential for parasite transmission to the mosquito.[334]
- Pf332 protein
The 700 kiloDalton protein Pf332 is the largest known exported asexual malaria protein. The protein has three parts: an N-terminal Duffy binding like domain followed by a putative transmembrane region and a large number of negatively charged repeats that are not identical but have the consensus (X)3-EE-(X)2-EE-(X)2–3 where E is glutamic acid and X is a hydrophobic amino acid. The repeat portion of the protein consititue more than 90% of the protein. The protein has a predicted isoelectric point (pI) of 3.8. It is known to associate with the erythrocyte plasma membrane.[335][336]
The Pf332 protein can first be detected within the parasite at 20–24 hours post invasion, after which it translocates across the parasitotopherous vacuole membrane into the host cell cytosol.[337] It is initially synthesised in the endoplasmic reticulum and eported to the host cytosol. From there it is trafficked as part of a multimeric protein complex to Maurer's clefts. It may interact with two chaperone proteins - PF14_0700 (a hypothetical protein with a J domain) and PFB0595w (a heat shock protein 40).[338] It is associated with the cytoplasmic side of Maurer's clefts in a peripheral manner throughout trophozoite maturation and schizogony.[339] In the clefts both the N and C-termini are localised to the erythocyte cytosol.[339] Export of Pf332 is sensitive to treatment with Brefeldin A[337] The export signal appears to be encoded in the N terminal domain[340]
It interacts with the erythrocyte cytoskeleton and binds actin.[223][341]
- PHIST family
The Plasmodium helical intersperse sub-telomeric family is a collection of 72 small exported proteins.[342]
- Other proteins
The merozoite specific thrombospondin related anonymous protein (MTRAP) is thought to be released from the micronemes during merozoite invasion and mediates motility and host cell invasion through an interaction with aldolase.[343] MTRAP is a highly extended bifunctional protein that binds to an erythrocyte receptor and the merozoite motor. MTRAP specific antibodies fail to inhibit parasite development in vitro.
Thrombospondin related apical membrane protein (PTRAMP) is a surface exposed protein whose function is currently unknown.[343]
The phosphoprotein P0 is surface exposed during the asexual erythrocytic stages and antibodies to this protein appear to be protective.[301] It is also present on the surface of the merozoites.
The 60S stalk ribosomal acidic protein P2 (gene PFC0400w) as well as forming part of the ribosome complex is surface exposed where it forms homo-tetramers.[301] This protein is exported to the erythrocyte surface 26-28 post invasion and persists there for 6–8 hours. Treatment with antiP2 antibodies causes mitotic arrest at the first nuclear division and disruption of the tubovesicular network which is set up during the trophozoite stages. Removal of the antibodies al lows the reformation of the tubovesicular network and mitotic division to continue.
The receptor for the attachment protein PfRh4 has been identified as complement receptor 1.[344]
The mature parasite-infected erythrocyte surface antigen (MESA) is exported to the erythrocyte cytoplasm where it binds to the N-terminal 30 kiloDalton domain of the erythrocyte protein 4.1R via a 19-residue sequence.[345] This sequence is also found in a number of other proteins in the parasite. Their role in remodeling of the erythrocyte are still under investigation.
The Ring-Infected Erythrocyte Surface Antigen (RESA/Pf155) protein appears to affect the mobility of the erythrocyte membrane.[346]
The proteins Pf12, Pf34, Pf92 and Pf38 are associated with detergent resistant membrane microdomains through glycosylphosphatidylinositol anchor sequences.[347] These microdomains are considered organizing centers for the assembly of molecules implicated in cell signaling.
The erythrocyte binding like 175 protein (EBL) is normally found in the micronemes. It has a signal sequence and a cysteine rich domain both of which are required for localisation to the micronene. In addition to these two ssequences for correct trafficking it also requires a sequence in its region 5.[348]
Enolase is bound to the surface of P. falciparum and several other pathogens.[349] In this location it binds plasminogen which is thought to function in the degradation of the extracellular matrix surrounding the targeted host cell, thereby facilitating pathogen invasion.
The gene PFE0565w is transcribed in both the erythrocytic and sporozoite stages.[350] The protein is only expressed in the salivary gland sporozoite stage.
Surfin 4.1 - a type I transmembrane protein located on the merozoite surface - is responsible for reversible adherence to the erythrocyte before invasion.[351] The gene is highly polymorphic, particularly at the C-terminal side of the variable region located just before a predicted transmembrane region. Positive diversifying selection is detectable in this region and in the conserved N-terminally located cysteine-rich domain.
Heparin has been shown to bind to infected erythrocytes.[352] What role this binding has in the pathology of this infection - if any - is not yet clear.
Erythrocyte proteins taken up
A small number of erythrocytic proteins are taken up by the parasite during the course of its life cycle. The role these play is not clear. Among these proteins is dematin which interacts with the parasite's 14-3-3 protein.[353]
The parasite is capable of making use of the erythrocyte's own enzymes. The enzymes PAK1 and MEK1 neither of which are encoded in the Plasmodium genome have been shown to be phosphorylated and activated during the course of infection[354]' In vitro work has shown that inhibition of these enzymes is fatal to the parasite.
Plasmodium ingests kininogen from which its proteases generate vasoactive peptides.[355] The role this may play in the pathophysiology of malaria is not yet understood.
Another enzyme that is imported by the parasite is the human redox-active protein peroxiredoxin 2 (hPrx-2, hTPx1).[356] This imported protein accounts for ~50% of the total thioredoxin peroxidase activity in parasite extracts. In the presence of chloroquine the parasite increases its imports of this protein. This protein is found both in the cytosol and in Maurer's clefts.
Another enzyme that is taken up is delta-aminolevulinate dehydrase - an enzyme in the haem biosynthesis pathway.[357]
Transport/secretion
The uninfected erythrocyte lacks a regulated transport system. Vesicular transport within both the parasite and the infected erythrocyte cytoplasm must be provided by the parasite itself.
Both the cytoplasmic pH (7.3) and the inside negative plasma membrane potential (-95 mV) are kept fairly constant during the intra erythrocytic cycle. This is due to the action of a V-type H(+)-ATPase which is also responsible for the pH of the digestive vacuole. There is also a Na+ ATPase in the plasma membrane.[358]
- Transport
The intracellular concentration of chloride ions has been estimated to be 48 milliMolar.[359] It appears to actively import using ATP both hydrogen ions and chloride ions in a linked fashion via a DIDS sensitive transporter in the cytoplasmic membrane.
One difficulty the parasite has in acquiring nutrients from the cytoplasm is the presence of phosphate groups on these molecules. It appears to have overcome this by secreting an acid phosphatase (glideosome-associated protein 50 - GAP50 ) into the cytoplasm that is then taken up into the digestive vacuole.[360]
The parasite has an absolute requirement for isoleucine - an amino acid absent from human haemoglobin. A saturable neutral amino acid (methionine, leucine, isoleucine) transporter appears to be encoded by the parasite and this protein functions in the infected erythrocyte membrane.[361]
The P. falciparum Na+/H+ exchanger (PfNHE1) is located on chromosome 13 (gene PF13_0019).[362] This gene may be involved in resistance to quinine.[363]
Two folate transporters (PfFT1 and PfTF2) have been cloned.[364] Substrates include folic acid, folinic acid, the folate precursor pABA and the human folate catabolite pABAG(n). 5-methyl tetrahydofolate is not transported by PfFT1 and only poorly by PfFT2. The activity of both transporters may be inhibited by probenecid or methotrexate. Folate transport appears to be an ATP requiring activity and dependent on a proton gradient.[365]
The parasite possesses its own equilibrative nucleoside transporter 1. All members of this protein family have 11 transmembrane segments. The gene product is located in the parasite's plasma membrane and knock out mutants have shown that this is an essential gene at least at physiological concentrations. In the 11th transmembrane segment two mutations have been shown to affect its activity: a phenylalanine (Phe) to leucine (Leu) at residue 394 (F394L) via cytosine (C) or uracil (U) to adenosine (A) or guanine (G) at the third codon position and a cysteine (Cys) to glycine (Gly) mutation at either glycine in a conserved glycine-X-X-glycine motif (where X is any amino acid) via a cytosine to uracil at the second codon position.[366] Additional work suggests that the 11th transmembrane segment is largely alpha helical. It has been suggested that this transmembrane segment may be the actual purine transport channel.
The parasite is unable to synthesize purines (including adenosine, hypoxanthine and adenine) and must take these up from the host. Purines are transported across the parasite plasma membrane entry into the infected erythrocyte P. falciparum nucleoside transporter 1 (PfNT1).[367] This transport system carries hypoxanthine, inosine and adenosine into the parasite. At least some of the hypoxanthine is converted into uric acid by the parasite.
Three purine transporters have been studied: the human equilibrative nucleoside transporter (hENT1), the human facilitative nucleobase transporter (hFNT1) and the parasite-induced new permeation pathway (NPP). The bulk of transport is facilitated by host's own transporters rather than through the NPP.[368] Hypoxanthine and adenine were transported mainly through the hFNT1 pathway whereas adenosine entered predominantly through the hENT1 system. The rate of purine uptake in infected cells was approximately twice that of uninfected erythrocytes. The rate of adenosine uptake was greater than the rate of hypoxanthine uptake in infected human red blood cells. Furosemide inhibits the transport of purine bases through the hFNT1.
An intracellular purine permease (PfNT2) has been shown to be localised to the endoplasmic reticulum.[369] This protein is a member of the equilibrative nucleoside transporter family.
Within the genome there are encoded four equilibrative nucleoside transporters (ENTs). ENT 1 is the major route of purine nucleoside/nucleobase transport in the erythrocytic stages. Knock out mutants have been generated that can survive. ENT4 has been cloned and expressed.[370] It does not appear to transport either hypoxanthine or adenine monophosphate but does transport adenine and 2'-deoxyadenosine. It is inhibited by dipyridamole.
The parasite can uptake polyamines from the host. Two of these - putrescine and spermidine - are taken up in a temperature, pH and membrane potential dependent mechanism.[371]
The clag3 genes on chromosome 3 appear to be involved in anion transport rather than in cell adherence as originally thought.[372]
At least two of the clag3 genes appear to be involved in the surface anion channel which functions in nutrient uptake.[373]
The clag3 gene family encode a parasite ion channel known as the plasmodial surface anion channel. Its activation appears to involve an intracellular domain.[374] The anion channel is formed by 5 clag genes and forms a complex with RhopH and other proteins.[375] The expression of the anion channel can be suppressed by reversible histone modification.[376]
Two paralogous clag3 genes - clag3.1 and clag3.2 - show mutually exclusive expression and can be silenced by epigenetic mechanisms.[377] They appear to offer different transport efficiency for some solutes. The expression of at least one of these genes is essential for the function of the anion channel.
Positive diversifying selection has acted upon clag2, clag8 and clag9 but not in clag3.1 and clag3.2.[378] These proteins appear to be involved in anion transport.
The plasma membrane protein aquaglyceroporin mediates the transport of both glycerol and water.[379]
A copper transport protein (PF14_0369) has been identified[380] This protein is expressed in early ring stage and translocating from the erythrocyte plasma membrane to a parasite membrane as the parasites developed to schizonts. Inhibition of copper uptake with neocuproine inhibits the ring to trophozoite transition.
The parasite is dependent on the acquisition of pantothenate from the host. The transporter - PfPAT - has been cloned.[381] The transporter is located in the parasite's plasma membrane and plays an essential role in its intraerythrocytic development. It can be inhibited by the drug fenpropimorph.
- Secretion
Protein export into the infected erythrocyte is critical for malaria parasite survival and the majority of effector proteins are thought to export via a proteinaceous translocon.[382] This is found in the parasitophorous vacuole membrane surrounding the parasite. Exported protein 2 is a critical component of this system.
A translocon (PTEX) of several proteins is located in the vacuole membrane.[383] These include heat shock protein 101 (HSP101) a ClpA/B-like ATPase from the AAA+ superfamily, of a type commonly associated with protein translocons, a novel protein termed PTEX150 and a known parasite protein - exported protein 2 (Exp2). Exp2 is the potential channel as it is the membrane-associated component of the core PTEX complex. Two other proteins - a new protein PTEX88 and thioredoxin 2 (Trx2) - are also PTEX components. These latter two proteins do not appear to be essential.[384] Deletion of either of these last two proteins is associated with a reduction in the replication rate in the blood.
PTEX88 is diffusely located within the blood stage parasites.[384] In trophozoites, PTEX88 is also localized to previously unrecognized extensions reaching from the parasite surface into the erythrocyte cytoplasm.
The thioredoxin 2 protein is part of the multi-protein complex embedded within the parasitophorous vacuolar membrane and is thought to be involved in protein secretion.[385] This protein is located in distinct punctate organelles of unknown identity.[384]
Within the genome are encoded 11 Rab GTPases.[386] These proteins are typically involved in vesicle transport. Casein kinase-1 has been shown to interact with RAB5B and the catalytic subunit of cAMP-dependent protein kinase A interacts with RAB5A and RAB7.
Several proteins are transported across its plasma membrane, the surrounding parasitophorous vacuole membrane and into its host erythrocyte. Most of these exported proteins contain a host targeting motif. Cleavage is of this motif by the protease plasmepsin V is normally part of this process. This process is not linked to the next component in the export pathway.[387] The fifth reside in the host motif is important for the action of plasmepsin V. Mutation of the fourth and fifth positions of this motif, as well as amino acids further downstream, block or affect the efficiency of protein export.
Surfin 4.1 is a type 1 transmembrane protein expressed on the surface of infected erythrocytes. It is exported to the surface via the endoplasmic reticulum/Golgi apperatus. Its structure varies between strains: in 3D7 there are 19 amino acids after the transmembrane region while FCR3 there are two tryptophan rich domains after the transmembrane domain.[225] The transmembrane region is required for the initial movement of the protein to the endoplasmic reticulum. The subsequent sorting step to the parasitophorous vacuole is determined by two independent signals located within the N terminual 50 amino acids. It may form a homodimer during transport.
Sodium is actively extruded from the parasite: at least one of the mechanisms involved in this process is PfATP4.[358]
A mutation in the mu-chain of the AP2 adaptor complex - a component of the endocytic machinery - has been associated with artemisinin resistance in Plasmodium chabaudi.[388] This protein interacts with a cargo recognition protein.
An essential protein - Cleft-like Protein 1 - has been cloned in P. berghei.[389] This protein has two predicted transmembrane domains in the C-terminal end. It is found in discrete convoluted, vesico-tubular membranous structures in the erythrocyte cytoplasm.
Band 4.1 is clover leaf shaped protein which interacts with multiple erythrocytic proteins via its three arms known as the N, C and alpha lobes. This protein is involved in maintaining the biconcave shape, elasticity, and mechanical stability of human erythrocytes, and defects in 4.1R are one cause of hereditary erythrocyte elliptocytosis. The mature parasite-infected erythrocyte surface antigen (MESA) interacts with the C lobe.[390] A secreted protein - PF3D7_0402000 - is localised to the parasitophorous vacuole membrane and interacts with Band 4.1.
Several of the exported proteins - PfEMP1, PfEMP3, ring associated erythrocyte surface antigen (RESA) and knob associated histidine rich protein (KAHRP) - interact with the preponderant erythrocyte skeleton protein spectrin.[391] KAHRP also binds to ankyrin R. KAHRP binds to ankyrin via a 79 residue segment: the reciprocal binding site for KAHRP is a subdomain (D3) of the ankyrin R membrane binding domain. KAHRP is normally associated with the host cell membrane. Blocking this interaction with ankyrin R prevents KAHRP movement to the host membrane: instead KAHRP remains diffusely distributed throughout the erythrocyte cytosol.
Kinases
Although several kinases are known in P. falciparum (~90 in total[392]) very little is known about them.
- Cyclin dependent kinases
A subgroup of cyclin-dependent kinases (CDK) including crk-5 have an activation loop that contains a novel Proline-Threonine-x-Cytosine motif which is absent from all known CDKs outside the Apicomplexa.[393]
The protein PFD0975w appears to be homologous with the right open reading frame 2 kinase RIO-2, a kinase involved in ribosome biogenesis and other cell cycle events.[394] This enzyme is unique among the kinases in the genome because along with the kinase domain, it also has a highly conserved N-terminal winged helix domain.[395]
The right open reading frame 2 protein kinase may be a potential drug target.[396]
- Cyclin dependent like kinases
Several are cyclin dependent like kinases (CLK): of these two - the Lammer kinase homologue PfCLK-1 and PfCLK-2 have been cloned.[397] CLKs in other eukaryotes are involved in the regulation of mRNA splicing through phosphorylation of serine/arginine-rich proteins. Both are transcribed throughout the asexual blood stages and in gametocytes. PfCLK-1/Lammer possesses two nuclear localization signal sites while PfCLK-2 possesses one of these signal sites upstream of the C-terminal catalytic domains. The two PfCLKs form complexes with proteins with predicted nuclease, phosphatase or helicase functions.
Although the kinases are primarily localized in the parasite nucleus, PfCLK-2 is also present in the cytoplasm. They are important for completion of the asexual replication cycle. Substrates phosphorylated by the PfCLKs include the Sky1p substrate, splicing factor Npl3p, and the plasmodial alternative splicing factor PfASF-1.
- NIMA kinases
Within the genome is a family of four protein kinases (Pfnek-1 to -4) that are related to the NIMA (never-in-mitosis/Aspergillus) family of kinases. The members of this latter family play important roles in mitosis and meiosis. Pfnek-1 (PFL1370w) is expressed in asexual parasites and male gametocytes.[398] It is an essential gene for completion of the asexual cycle. The other three - Pfnek-2 (PFE1290w), -3 (PFL0080c) and -4 (MAL7P1.100) - are expressed predominantly in gametocytes.
Pfnek-2 is predominantly expressed in gametocytes and is required for DNA replication during meiosis and ookinete development.[190]
The plasmodial mitogen-activated protein kinase kinase Pfnek-3 has both serine/threonine and tyrosine kinase activities.[399]
Pfnek-4 is expressed in stage II to V gametocytes and in a subset of asexual stage parasites undergoing schizogony.[400] It is also required for the completion of meiosis in the ookinete.[190]
- Adenylate kinases
There are at least three adenylate kinases (AK) encoded in the genome - PfAK1, PfAK2 and a GTP:AMP phosphotransferase (PfGAK).[401] There are two additional adenylate kinase-like proteins - PfAKLP1 (which is homologous to human AK6) and PfAKLP2. PfAK1, PfAKLP1, and PfAKLP2 are found in the cytosol. PfGAK is located in the mitochondrion. PfAK2 is located at the parasitophorous vacuole membrane and this localization is driven by N-myristoylation.
Adenylate kinases are phosphotransferases that catalyze the interconversion of adenine nucleotides. There are at least three adenylate kinases (PfAK1, PfAK2 and GTP:AMP phosphotransferase) encoded in the genome.[402] PfAK1 and PfAK2 both catalyse the conversion of ATP and AMP to two molecules of ADP. PfGAK instead has a preference for GTP and AMP and does not accept ATP as a substrate.
- Calcium dependent kinases
The calcium dependent protein kinases (CDPK) are part of a superfamily found in plants, ciliates and some apicomplexa. They are not present in fungi or animals. They have three domains: a variable N-terminal region involved in substrate recognition and protein interaction, a kinase catalytic domain and a regulatory domain. The regulatory domain has two subdomains - an autoinhibitory junction domain and a calmodulin like domain. The calmodulin domain has four EF hands. These hands, upon binding calcium, undergo a structural change that moves the junction domain from its autoinhibitory interaction with the substrate binding site of the kinase domain which in turn activates kinase domain catalytic activity.
Seven CDPKs are present in the genome.[403] The first of these cloned was PfCDPK2 in 1997.
Calcium dependent protein kinase 1 is expressed in parasite asexual blood and mosquito stages.[404] This protein has a specific auto inhibitory junction region (J). It localises to the parasite plasma membrane of very young intracellular parasites, replicating and invasive forms. It does not appear to be exported into the erythrocyte. Inhibition of this protein results in the arrest of parasite development late in the cell cycle during early schizogony. This protein also appears to have a role in microneme secretion during the process of merozoite invasion of the erythrocyte.[405] It also phosphorylates members of the actin-myosin complex.[406]
Calcium dependent protein kinase 1 autophosphorylates at several site and has an ATP binding site in its N terminal.[407] The protein is myristoylated at its N terminus and is localised to the parasitophorous vacuole and the tubovesicular system of the parasite.[408] It is also palmitoylated at its N terminus and has a basic motif located there.
The homolog of calcium dependent protein kinase 1 (CDPK1) in Toxoplasma gondii is calcium dependent protein kinase 3 (TgCDPK3). This protein in Toxoplasma is localised to the inner membrane and is not an essential gene.[409] It is involved in Ca(2+) ionophore control and host cell egress. The role of this protein in Plasmodium is not currently known. It is however expressed and localises with proteins at the periphery of the schizonts and merozoites involved in gliding motility[410] and can can phosphorylate these proteins.[411] Inhibition of CDPK 1 is associated with a block in development at the schizont level.[410] In P bergei CDPK1 regulates transcription of stored mRNA during ookinete development in the mosquito midgut.[412]
In P. falciparum CDPK5 controls parasite egress from host cells.[413] In P. bergei CDPK3 is essential for the ookinete to traverse the mosquito midgut epithelium[414] and CDPK4 is involved in development of the male gametocyte.[415] In P. falciparum CDPK4 appears to be involved in the exflagelation of the male gametes.[416]
- Mitogen activated kinases
A mitogen activated protein kinase (MAP kinase) gene is located on is located on chromosome 14.[417] It is predominantly expressed in gametocytes and gametes/zygotes. The protein has 882 amino acid residues and possesses a TDY dual phosphorylation site upstream of the highly conserved VATRWYRAPE sequence within subdomain VIII. Within the carboxyl-terminal segment the protein contains an unusually large and highly charged domain. This region includes two repetitive sequences of either a tetrapeptide or octapeptide motif.
Two mitogen activated protein kinases are present in the genome.[418] Both genes appear to be transcribed during the liver stages.
An atypical mitogen activated protein kinase (MAPK) - Pfmap-2 - is known.[419] It posses the usual properties of a MAPK - including (i) the ability to undergo autophosphorylation, (ii) the ability to phosphorylate myelin basic protein, a classical MAPK substrate, (iii) the regulation of kinase activity by a MAPK-specific phosphatase and (iv) the ability to be activated by component(s) present in cell extracts. It is expressed in gametocytes. It lacks the conserved threonine-X-tyrosine activation motif usually found in enzymes of this family and instead has a threonine-serine-histidine at the same location.
- FIKK kinases
A groups of 20 kinases with a Phe-Ile-Lys-Lys sequence motif appear to be unique to P. falciparum.[420] One of these kinases (PfFk4.1, PFD1165w) has been cloned. It autophosphoralates and phosphorlyates dematin, a cytoskeletal protein found at the erythrocyte spectrin-actin junction.
- Other kinases
The protein kinase CK2, a serine/threonine protein kinase, has one catalytic subunit (PfCK2) and two regulatory ones (PfCK2beta1 and PfCK2beta2).[421] This enzyme is found both in the cytoplasm and the nucleus. Substrates include the nucleosome assembly proteins (Naps), histones and two members of the Alba family. Both of the two regulatory subunits are required for completion of the asexual erythrocytic cycle.
The cyclic guanine monophosphate dependent protein kinase is essential for the initiation of gametogenesis and for blood stage schizont rupture and may also be involved in ookinete differentiation and motility and liver stage schizont development.[422]
Pantothenate kinase, the first enzyme involved in converting pantothenate to coenzyme A is present in the genome.[423] It appears to be an essential enzyme.
A putative O-phosphoseryl-tRNA(Sec) kinase - an enzyme involved in the formation of selenocysteine tRNA - has been identified in the genome.[424] A protein kinase C has been identified.[425] Although predominantly cytosolic it is also present in the membrane faction. Its activation requires Ca2+, phosphatidyl serine and either diacylglycerol or phorbol myristate acetate. Its activity is 9 time greater in the trophozoites than in the ring forms. On activiation in the trophozoites the activity in the membrane farction increase significantly. Ita actvity is inhibited in a dose dependent fashion by chloroquine. The inhibition appears to be non compeditive. Chloroquine resistant strains do not show this inhibition of activity.
A tyrosine kinase like kinase (PfTKL2) of the IRAK/RLK/Pelle protein family has been identified in the genome.[426] The gene is expressed in asexual blood stages and in gametocytes. It is also secreted into the culture media. Its function is not presently known.
Another kinase - PfPK7 - is involved in the rate of asexual growth in erythrocytes, the production merozoites in the schizonts and in the production of oocysts in the mosquito vector.[427]
Phosphatases
There are 27 putative protein phosphatases in the genome. These can be classed into groups: phosphoprotein phosphatases, metallo-dependent protein phosphatases, protein tyrosine phosphatases and NLI interacting factor-like phosphatases.[428]
A Shewanella-like protein phosphatase that is expressed at all stages of the parasite life cycle but is particularly bundant in asexual blood stages and expressed at all stages of the parasite life cycle is essential for ookinete (zygote) development and microneme formation.[429]
The open reading frame PF3D7_1305500 encodes an atypical MAPK phosphatase. This gene appears to be involved in the regulation of the transition from the pre-S phase to the M phases of asexual intraerythrocytic development.[430]
Protein phosphatase 1 is a key enzyme that plays diverse and essential roles in cell biology. Its dephosphorylation activity/specificity is governed by the interaction of its catalytic subunit (PP1c) with regulatory proteins. A homolog of the inhibitor of protein phosphatase 1 has been cloned.[431] This gene is essential for survival and appears to be localised to the nucleus. A conserved 41- Lysine-Valine-Valine-Arginine-Tryptophan-45 motif is essential for its inhibition activity. Another inhibitor - inhibitor 2 - has also been cloned.[432] Within the protein are two motifs - 12-Lysine-Threonine-Isoleucine-Serine-Tryptophan-16 and 102-Histadine-Tyrosine-Asparagine-Glutamine-105 - that are critical for its activity. It appears to be an essential gene and seems likely to act at the G2/M point of the cell cycle.
Cysteine proteases
A number of cysteine proteases have been identified this organism including four falcipains, serine repeat antigens (SERA), dipeptidyl aminopeptidase 1, dipeptidyl aminopeptidase 3 and a calpain homolog.[433]
- Falcipains
The falcipains belong to the papain family of enzymes (clan CA).
Falcipain-1 appears to be important in the development of the oocysts in the mosquito. The ortholog of this protein in Plasmodium chabaudi - chabaupain-1 - is localised preferentially to the apical portion of the ookinete.[434]
Falcipain-2 is involved in the hydrolysis of haemoglobin and appears to be a non essential gene. It also promotes host cell rupture by cleaving the skeletal proteins of the erythrocyte membrane.
Falcipain-3 appears to be an essential gene but its function has yet to be firmly established.
- Dipeptidyl aminopeptidases
Dipeptidyl aminopeptidase 1 is found in the digestive vacuole and is also an essential gene.
Dipeptidyl aminopeptidase 2 is specific to the gametocyte. It does not appear to be an essential gene.[435]
Dipeptidyl aminopeptidase 3 appears to be involved in the release of the merozoites from the erythrocyte.
- Serine repeat antigens
The serine repeat antigens are a set of nine proteins in P. falciparum (two in P. gallinaceum, five in P. bergei, 12 in P. vivax) with a central domain homologous to the papain-like (clan CA, family C1) protease family.[164] They are named after the presence of tandem repeats of serine residues in the protein.
The only known homolog of the SERA proteases is found in the parasite Theileria.[436] In a study of 18 species the majority of the SERA genes were found to be clustered between between two conserved genes: a conserved hypothetical protein (HP) gene and the iron-sulfur assembly protein gene (hesB). There is a signal peptide sequence at the N-terminus. The family can be divided into four groups: Groups I to III have a cysteine at the active site and Group IV has a serine residue. Group II and III are sister clades and are more closely related to Group I than to Group IV. With a few exceptions the SERA gene family has a common structure of four exons and three introns. SERA genes of Group I in general have a six exon and five intron structure. In the mammalian malaria parasites, gene duplication occurred only in Group IV SERA genes and was particularly frequent in primate species. The reasons are unknown presently.
Although most SERAs are cysteine proteases the majority in P. falciparum (6 of the 9) have a serine residue at the active site. The SERAs possessing a canonical Cys residue at the position of the active site nucleophile in their papain-like central domains are with SERA6, 7 and 8.
SERA5 and SERA6 are indispensable in blood-stage parasites. SERA 5 (molecular weight 120 kiloDaltons) is abundantly produced at the late trophozoite to schizont stages of parasite development. SERA 6 is produced in much smaller quantities than SERA 5 and appears to be involved with egress from the erythrocyte. SERA 6 is activated by SUB1 - a seine protease that is stored in exonomes.
SERA8 in P. falciparum (SERA5 in P. berghei) is expressed exclusively during mosquito stages of the parasite life cycle, where it has been shown to be required for egress of midgut sporozoites from oocysts.[186]
- Other proteases
A calpain homolog has been cloned.[437] Although shorter than other calpains it possesses a typical catalytic triad (Cysteine-Histamine-Asparagine).[438]
A cysteine protease inhibitor - falstatin - is involved in regulating proteolysis during erythrocyte infection.[439] It is found in vesicles within the asexual blood stage parasite cytoplasm, the parasitophorous vacuole and is exported to dynamic exomembrane structures in the infected erythrocyte. In sporozoites, its is found in the rhoptries and in intracellular vesicles distinct from the micronemes. In the final stages of liver infection it is released into the infected hepatocyte during parasitophorous vacuole membrane breakdown.
Homologs of these proteins have been identified and cloned in Plasmodium knowlesi.[440] These three knowpains are found in the digestive vacuole, are active at acidic pH and are capable of degrading haemoglobin. Two of them (KP2 and KP3) cleave only if the P2 position is occupied by an leucine residue. KP4 shows a moderate preference for leucine at the P2 position but is more active if this position is occupied by an arginine residue. Although found in the digestive vacuole KP4 is also found in the parasite periphery and may play a role in parasite egress from the erythrocyte.
Metallopeptidases
There is at least one M1 family aminopeptidase in the genome (PfA-M1). This is a zinc binding metalopeptidase with optimal activity at pH 7.4, and remains at least 40% active between pH 5.8-8.6. It is an alanyl aminopeptidase. Immunofluorescence studies have shown that in trophozoites that it diffusely found in the parasite cytoplasm with accumulations outside the digestive vacuole while in schizonts it is progressively located to a vesicle like pattern ending as a single location in released merozoites. It exists as two major isoforms, a nuclear 120 kDa species and a processed species consisting of a complex of 68 and 35 kDa fragments.[441]
There are at least 2 essential metallopeptidases encoded in the genome - PfA-M1 and Pf-LAP.[442] Specific inhibition of PfA-M1 causes swelling of the parasite digestive vacuole and prevented proteolysis of haemoglobin derived oligopeptides. This inhibition is lethal to the parasite probably by starvation.
Leucyl aminopeptidase (LAP) is a member of the M17 family. It has a predicted molecular weight of 67.831 kiloDaltons and is localized in the cytosol. It is inhibited by bestatin. Its inhibition is lethal to parasites early in the life cycle, prior to the onset of haemoglobin degradation suggesting a different role for this enzyme.
Falcilysin a zinc metalloprotease found in the apicoplast.[253] It is a member of the M16 protease group and has maximal activity at neutral pH. It appears to be an essential gene. Its function in this organelle is not quite clear but it appears to be involved in the degradation of transit peptides.
M18 AAP is a metallo-aminopeptidase that has a highly restricted specificity for peptides with an N-terminal glutamine or asparagine residue.[443]
The gene PFI1625c appears to be a metaloprotease.[444]
Aspartyl proteases
At least four aspartyl proteases known as plasmepsins are involved in the degradation of haemoglobin by Plasmodium falciparum.[445]
The histo-aspartic protease (HAP) has been crystallised.[446] This protein has high sequence similarity to pepsin-like aspartic proteases, but one of the two catalytic aspartates, Asp32, is replaced in this enzyme by a histidine residue. The propeptide interacts with the C-terminal domain of the enzyme, forcing the N- and C- terminal domains apart. This mechanically separates His32 and Asp215 and prevents formation of the mature active site. This mechanism is similar to those of other proplasmepsins. The enzyme has a number of unique features and may be a useful drug target.
There are at least 10 aspartic proteases encoded within the genome. Plasmepsins I, II, IV and histo-aspartic protease are known to be involved in the digestion of haemoglobin.[133] These four enzymes share 50-79% amino acid sequence identity[447] and are located on chromosome 14 (gene identifiers PF14_0076, PF14_0077, PF14_0078, and PF14_0075 respectively).[448] Plasmepsins I and II are present in the food vacuole and make the initial cleavages in the hemoglobin molecule. The proplasmepsins I and II are both type II integral membrane proteins that are transported through the secretory pathway before cleavage to the soluble form. This reaction occurs within the food vacuole and the cleavage occurs immediately after a conserved Leucine-Glycine dipeptidyl motif.[449] This reaction may be blocked calpain inhibitors. It appears that plasmepsin II and IV are capable of autoactivation as well as activation each other's inactive form.[450] These two proteins are not glycosylated. Plasmepsin I is synthesized and processed to the mature form soon after the parasite invades the red blood cell, while plasmepsin II synthesis is delayed until later in development.
- Intramembrane proteases
Plasmepsin V, an integral membrane protein, is located within the endoplasmic reticulum but not in the Golgi apperatus.[451] The gene is expressed over the course of asexual intraerythrocytic development. The amount of the protein in the parasite is lowest in the ring stage and increases steadily through schizogony. It appears to be involved in the export of proteins to the erythrocyte. Plasmepsin V cleaves N-terminal sequences from RIFIN, STEVOR and RESA multigene families but does not cleave the N-terminal sequence of erythrocyte membrane protein 1, skeleton binding protein (SBP-1) or REX-2.[452]
A presenilin-like signal peptide peptidase is known to be present in the endoplasmic reticulum.[453]
Serine proteases
A SUMO specific protease PfSENP1 (PFL1635w) has been identified in the genome but its importance if any is not known[454]
There are at least 3 subtilisin like proteases encoded in the genome.[455] These are serine proteases.
SUB1 has multiple roles in the parasite's life cycle: in the blood stages it is involved in the egress of merozoites from the infected erythrocytes and also in priming merozoites for subsequent erythrocyte invasion.[456] It also appears to be involved in escaping from the parasitophorous vacuole membrane in the hepatocytes. Within the merozoites SUB1 is stored in exonemes[457] and released in response to a rise in calcium.[458] Blocking this release results in failure to process serine repeat antigen 5 (PfSERA5) and parasitophorous vacuolar membrane rupture and merozoite egress from the hepatocytes. SUB1 also acts on merozoite surface protein 1 (MSP1), MSP6 and MSP7 on the merozoite surface.[459] Blocking the action of SUB1 on these proteins reduces the ability of the merozoite to invade erythrocytes. SUB1 also acts on serine repeat antigen 6 (SERA 6) - a cysteine protease found in the intraerythrocytic parasitophorous vacuole.[164] The action of SUB1 activates the proteolytic function of SERA 6.
In the merozoites SUB2 has been implicated in shedding of adhesins at a juxtamembrane position.[460] PfSUB2 is located on the surface of the parasite.[461] Posttranslationally it is transported first to the micronemes and then to the surface. Its usual promoter is required for its final location. Its transmembrane domain is required for transport to the microneme and the cytoplasmic domain is required for its surface localization.
Another protease - PfSUB3 - is expressed at late asexual blood stages. This enzyme is known to act on profilin (PfPRF).[462]
- Intramembrane proteases
At least two rhomboid proteases (ROM1 and ROM4) are present in the genome.[306] They are able to act on a number of substrates including TRAP, CTRP, MTRAP, PFF0800c, EBA-175, BAEBL, JESEBL, MAEBL, AMA1, Rh1, Rh2a, Rh2b and Rh4 but not PTRAMP.[463]
The protease rhomboid-1 (ROM1: PF11_0150) is located within a single, thread-like structure on one side of the merozoites that appears to be in close proximity to the subpellicular microtubules. This organelle has been named the mononeme.[464] The mononeme first appears in the schizonts. On the release of merozoites from schizonts, ROM1 moves from a lateral asymmetric localization to the merozoite apical pole and the posterior pole. It can cleave the apical membrane antigen 1.[463]
ROM4 (PFE0340c) appears to be involved in the gliding motility of the sporozoite.
Actin
Within the genome are encoded two forms of the protein actin - I and II. The first form (I) is present in significantly greater quantities. Actin II appears to be essential for the process of exflagellation.[465] Deletion of this gene results in viable asexual stages. During the formation of the male gametes actin I is found initially in both the nucleus and the cytoplasm. After activation it is found only in the cytoplasm. In actin II deletion mutants actin I remains in both the nucleus and the cytoplasm after activation. Morphologically in the actin II mutants male gametocyte DNA was replicates normally and axonemes are assembled but egress from the host cell is inhibited and axoneme motility is abolished.
Two proteins P. falciparum actin-depolymerizing factor 1 (PfADF1) and P. falciparum actin-depolymerizing factor 2 (PfADF2) are involved in the polymerisation of actin.[466] PfADF1 has ben crystallised and despite having significant differences from other proteins with similar function it is capable of severing actin filaments. PfADF2, like canonical ADF proteins but unlike ADF1, binds to both globular and filamentous actin, severing the filaments and inducing nucleotide exchange on the actin monomer.[467] The crystal structure of PfADF1 shows major differences from the ADF consensus, explaining the lack of F-actin binding. PfADF2 structurally resembles the canonical members of the ADF/cofilin family.
The actins found In Plasmodium and in Toxoplasma are divergent both in sequence and function and only form short, unstable filaments in contrast to the stability of conventional actin filaments.[468] This inherent instability of parasite's actin filaments is a critical adaptation for their gliding motility.
Actin is involved in the expression of the var genes.[11] The var introns interact with an 18 base pair nuclear protein binding element which recruits actin and repositions the var DNA from a transcriptionally repressive to a transcriptionally active perinuclear compartment.
The presence of actin microfilaments has been demonstrated in the ookinete in the pellicle and in the apices.[469]
There are two formin genes encoded in the genome.[470] These associate with and nucleate both mammalian and Plasmodium actin filaments. Another gene profilin - also encoded in the genome but only as a single copy - sequesters actin monomers preventing their polymerisation.
Aldolase, an actin binding protein, is involved in the moving junction that forms during the invasion of the erythrocyte.[343]
Several hemoglobinopathies that protect carriers from severe malaria may do so by interfering with host actin reorganization.[220]
The cyclase associated proteins are among the most highly conserved regulators of actin dynamics. They catalyze nucleotide exchange on actin monomers from ADP to ATP and recycle actin monomers from ADF/cofilin for new rounds of filament assembly. The Plasmodium falciparum cyclase associated protein is entirely composed of β-sheet domains and efficiently promotes nucleotide exchange on actin monomers.[471]
Ubiquitin
The addition of the small protein ubiquitin to other proteins as part of post translational processing is widespread in most eukaryotes. This is also the case with P. falciparum where this process occurs at all stages of the asexual life cycle.[472] Ubiquitylation involves the covalent attachment of a ubiquitin moiety to lysine residues of protein substrates via the hierarchical intervention of an E1 ubiquitin activating enzyme, an E2 ubiquitin conjugating enzyme, and an E3 ubiquitin ligase that is usually involved in specific substrate recognition. This process is also known as sumoylation (small ubiquitin related modifiers).
Ubiquitylation is involved in removing misfolded proteins from the endoplasmic reticulum - a process known as Endoplasmic-reticulum-associated protein degradation. This process is a prerequisite for subsequent retro-translocation to the cytosol and destruction by the 26S proteasome. Aberrant proteins are recognized by endoplasmic reticulum luminal chaperone proteins and protein disulfide isomerases to help discriminate properly folded proteins from misfolded proteins. Misfolded proteins are shuttled to the DER1 translocon complex which forms a hydrophobic pore to allow the retro-translocation of proteins through the endoplasmic reticulum membrane. Several components of the system are known to be present in the parasite: HRD1 (E3 ubiquitin ligase), UBC (E2 ubiquitin conjugating enzyme) and UBA1 (E1 ubiquitin activating enzyme).[294] HRD1 localizes to the endoplasmic reticulum membranes, while UBC and UBA1 localize to the cytosol. HRD1 interacts with membrane bound proteins needed for retro-translocation and helps form the hydrophobic pore complex. Another member of this pathway is the signal peptide peptidase.
The enzymatic mechanism is reasonably well understood. The E1 enzyme adenylates ubiquitin at its C-terminus, creating a mixed anhydride: this process requires ATP. The sulfhydryl group of the E1 active site cysteine then attacks the anhydride: thus results in the formation of a high energy thio-ester linking ubiquitin to E1. Ubiquitin is then passed to the active site cysteine of the E2 enzyme. Finally, with the aid of an E3 ligase, ubiquitin is transferred from E2 and covalently attached to the ε-amino group of a lysine in the target protein.
The genes PFL1245w is the E1 ubiquitin activating enzyme, PFL0190w is the E2 ubiquitin conjugating enzyme and PF14_0215 is the E3 ubiquitin ligase. PFL1245w (E1) contains a ubiquitin activating enzyme active site, two ubiquitin like activating enzyme catalytic domains, two ThiF repeats and a catalytic cysteine at the N-terminal end. PFL0190w (E2) is 147 amino acid residues in length and contains an ubiquitin conjugating enzyme domain takes up almost its whole length. PF14_0215 (E3) has multiple transmembrane domains, an E3 RING zinc finger (zf-C3HC4) domain on its C-terminal half and a predicted signal peptide consistent with endoplasmic reticulum targeting. The presence of four transmembrane domains is compatible with a pore forming ability and to be able to participate in the recognition and translocation of misfolded proteins across the endoplasmic reticulum membrane.
The addition of ubiquitin to a protein frequently precceds its digestion in the proteasome. The proteasome mediates the nonlysosomal degradation of cytosolic proteins in eukaryotic cells. It is a large complex consisting of two multisubunit structures, the 20S and 19S (PA700) or P28 complexes which combine to form the 26S particles. The proteasome subunit 4 ATPase has been cloned from P. falciparum.[473] The protein is similar to those found in eukaryotes. It lacks introns.
The E1 and E2 enzymes interact to recognize and modify RanGAP1.[474] Sumoylation actvity peaks during mid stages of the intra-erythrocyte developmental cycle.
A mutation in a deubiquitinating enzyme in Plasmodium chabaudi has been reported to cause both artesunate and chloroquine resistance.[475]
It appears that ubiquitylation may be a mechanistic requirement of apicoplast protein import independent of the proteasomal degradation pathway.[476]
Heat shock proteins
A number of heat shock proteins 40 (hsp40) have been predicted from the sequenced genome. Only one is predicted to be a cytosolic canonical Hsp40 capable of interacting with the major cytosolic Hsp70 an interaction that has been confirmed experimentally.[477]
Heat shock protein 20 has been shown to have a critical role in sporozoite motility.[478] This role appears to be via substrate adhesion.
PfGECO is a type IV heat shock protein 40 expressed in gametocyte stages I to IV and is exported to the erythrocyte cytoplasm.[479] This gene appears to be non essential.
A Hsp40 class of chaperone (PFB0090c; PF3D7_0201800; KAHsp40) is located in a chromosomal cluster together with knob components KAHRP and PfEMP3.[480] This protein has a PEXEL motif required for transport to the erythrocyte compartment. It occurs in punctuate spots in the erythrocyte periphery, distinctly from Maurer's clefts. These structures may be knobs particularly since it is found in a complex the known knob proteins KAHRP, PfEMP3 and Hsp101.
There are 6 HSP70 proteins in the genome.[481] Five of these proteins are found in other species: the exception being PfHsp70-x.
Heat shock proteins Hsp70 and Hsp90 are both expressed in P. falciparum. They are linked by an essential adaptor protein known as the Hsp70-Hsp90 organising protein (Hop). This protein co-localises with PfHsp70 and PfHsp90 at the trophozoite stage and forms a complex with them.[482]
A heat shock protein PfHsp70-x with endoplasmic reticulum signal peptide has been identified.[483] It is maximally expressed at the schizont stage of intra erythrocytic life cycle. Although the majority of the protein localizes to the parasitophorous vacuole, some of it gets exported to the erythrocyte compartment where it associates with Maurer's clefts. It lacks an endoplasmic reticumlum signal and interacts with at least some of the HSP40 proteins.
The protein Aha1 interacts with HSP90.[484]
The heat shock protein 110 (Hsp110) is an essential protein in P. falciparum.[485] Although its usual function is still under investigation it is known to prevent the aggregation of asparagine repeat rich proteins within the parasite. These repeat rich proteins tend to form aggregates particularly at the elevated temperatures that occur with malaria.
A 20 kiloDalton protein (cpn20) that acts as a heat shock protein and is localised in the apicoplast has been cloned.[486]
Redox balance
The infected host cell is under considerable oxidative stress. Normal erythrocytes have a ratio of reduced (GSH) and oxidized glutathione (GSSG) of 321.6 while the GSH/GSSG ratio in infected cells is 26.7.[487] The ratio in the parasite is 284.5. Efflux of GSSG from the intact infected cell is more than 60-fold higher than the rate observed in normal erythrocytes. This export process is mediated by permeability pathways that the parasite induces in the erythrocyte's membrane. Exogenous gamma-glutamylcysteine is not converted into GSH in the infected erythrocyte suggesting that the erythrocytes' own GSH synthetase may not be functional. This may be due to the lower levels of magnesium (Mg2+) in the infected erythrocyte (0.5 milliMolar) compared to the normal erythrocytes (1.5-3 mM). The lower level of results in cessation of gamma-glutamylcysteine synthesis and of GSH synthesis in the infected erythrocyte. The parasite maintains a level of 4 mM magnesium. The parasite membrane is impermeable to both gamma-glutamylcysteine and GSH.
The proteins gamma-glutamylcysteine synthetase and glutathione synthetase which are involved in the synthesis of glutathione appear to be essential genes in P. falciparum.[488] Inhibition of the glutathione biosynthesis by the parasite is lethal. Its levels appear to be tightly regulated. The enzyme glutathione reductase is highly specific for its substrate glutathione disulfide.[489]
Glutathione export from parasitized cells is inhibited partially by both the compound MK571 and by furosemide.[490] These agents are inhibitors of the 'new permeability pathways' induced by the parasite in the host erythrocyte membrane.
A possible connextion between the redox system and chloroquine resistance has been described.[491] Strains carrying the mutant chloroquine resistant transport gene have lower levels of glutathione and are more sensitive to inhibition of glutathione synthesis. The mutant chloroquine resistance transport gene selectively transports glutathione into the digestive vacuole. Also in the mutant strains the haemozoin levels are lower and there is reduced chloroquine binding by haemozoin. It seems likely that the increased transport of glutathione into the digestive vacuole may alter both the synthesis of haemozoin within the digestive vacuole and the binding of chloroquine to haemozoin within the digestive vacuole.
The thioredoxin system like the glutathione system is responsible for maintaining the redox balance in the cell. The thioredoxin reductase reduces thioredoxin and a number of other low molecular weight compounds.[489] The other members of this system include five peroxiredoxins differentially located in the cytosol, apicoplast, mitochondria and nucleus with partially overlapping substrate preferences. It also includes members of the thioredoxin superfamily with three thioredoxins, two thioredoxin-like proteins, a dithiol and three monocysteine glutaredoxins and a redox-active plasmoredoxin being encoded in the genome.
Within the cytoplasm two peroxiredoxins - T peroxiredoxin-1 (TPx-1) and 1-Cys peroxiredoxin (1-Cys Prx) - are produced at differing points in the life cycle.[492] Disruption of the T peroxiredoxin-1 enzymes renders the parasite hypersensitive to heat stress. This does not occur with knock out mutants of 1-Cys peroxiredoxin suggesting that these enzymes have different roles in the life cycle.
Investigation of the liver stages of these enzymes in Plasmodium berghei has shown that both TPx-1 and 1-Cys Prx are present in the cytosol but differ in their expression patterns.[493] TPx-1 is transcribed shortly after infection of the hepatocyte and expression continues until the schizont stage. Transcription of 1-Cys Prx starts after the parasite has developed into the schizont stage.
The 1-Cys peroxiredoxin enzyme appears to be located in the cytoplasm.[494] The peroxiredoxin requires both glutaredoxin and glutathione for its activity.[495]
The crystal structure of thioredoxin reductase has been solved.[496] There are significant differences between the human enzyme and the plasmodial which are most apparent in the Plasmodium-specific insertion and the conformation of the flexible C-terminal arm.
Lysis of the erythrocyte releases methaemoglobin. Exposure of uninfected erythrocytes to methaemoglobin renders them susceptible to osmotic stress and haemolysis.[49] This mechanism may form part of a chain reaction resulting in the haemolysis that is found in severe malaria. Both N-acetyl cysteine and mannitol can reduce this oxidative stress. Clotrimazole irreversibly inactivates methaemoglobin and abolishes its peroxidase activity.
Ferriprotoporphyrin IX is released inside the digestive vacuole of the malaria parasite during the digestion of host cell hemoglobin.[497] Undegraded ferriprotoporphyrin IX accumulates in the membrane fraction and is degraded by reduced glutathione in a radical mediated mechanism.
Three phosducin-like proteins have been identified in Plasmodium berghei.[498] Their role in the parasite's metabolism has yet to be clearly established.
The type II NADH:ubiquinone oxidoreductase has been shown to be redundant in the blood forms but to be essential in the mosquito midgut.[499]
At least one function of B6 in this parasite is as an antioxidant.[500]
The thioredoxin like protein TRX2 is not essential but its deletion reduces growth rate.[501]
The glyoxalase system is the main catabolic route for methylglyoxal, a non-enzymatic glycolytic byproduct with toxic and mutagenic effects.[502] In Plasmodium falciparum the glyoxalase pathway is glutathione dependent. Its glyoxalase I is an atypical monomeric enzyme with two active sites.
Calcium fluctuation
In recently invaded erythrocytes the Ca2+ concentration increases about 10 fold.[503] The Ca2+ content increases as the parasite matures.[504] In infected erythrocytes, Ca2+ is almost exclusively localized in the parasite compartment and changes but little in the cytosol of the host cell.
Cytosolic calcium2+ increases evoked by extracellular stimuli are may be observed in the form of oscillating Ca2+ spikes in eukaryotic cells. Spontaneous spikes in the calcium levels have been observed in Plasmodium falciparum.[505] The frequency of Ca2+ oscillations are higher in early ring forms than that in early trophozoites. Blockage of this oscillation results in the cessation of intraerythrocytic maturation and death of the parasite. This effect is maximal in the trophozoites.
An inositol phosphate kinase with a role in calcium metabolism has been cloned.[506]
Expression of the calmodulin gene is developmentally regulated throughout the blood-stage cycle.[507]
Phosphoinositide-specific phospholipase C (PI-PLC) is a major regulator of calcium-dependent signal transduction, usually by liberation of calcium from intracellular stores through the action of its product, inositol-(1,4,5)-trisphosphate. These genes are found in P. falciparum and appear to be essential.[508] The genes are twice as long as their mammalian counterparts and belong to the delta class of phospholipase C proteins.
Nucleotide metabolism
P. falciparum is unable to biosynthesize purines.[3] Instead, the parasite is able to transport and interconvert host purines. The enzyme hypoxanthine-guanine-xanthine phosphoribosyltransferase converts hypoxanthine to inosine monophosphate and is essential for purine salvage.[509]
Conversely, the parasite can produce pyrimidines de novo using glutamine, bicarbonate and aspartate.[3]
A gene encoding S-adenosyl-L-homocysteine hydrolase is present in the genome.[435]
P. falciparum contains both cytosolic and mitochondrial serine hydroxymethyltransferase isoforms.[510] This is a pyridoxal phosphate dependent enzyme which plays a vital role in the de novo pyrimidine biosynthesis pathway. Both genes are expressed throughout the erythrocytic stages.[511] Both enzymes appear to be essential.
A conserved trytophan residue is involved in the activity of the purine nucleoside phosphorylase enzyme.[512]
Orotate phosphoribosyltransferase catalyzes the magnesium dependent condensation of orotic acid with 5-α-D-phosphorylribose 1-diphosphate to yield diphosphate and the nucleotide orotidine 5'-monophosphate. This enzyme has been crystallised.[513]
Molecular biology
The long adenosine/thymidine tracts that are scattered through the genome may play a role in gene duplication.[514]
The centromeres occupy a 4-4.5 kilobase region in each chromosome.[515] The centromeres cluster to a single nuclear location prior to and during mitosis and cytokinesis but dissociate soon after invasion.
The DNA polymerase is unusual.[516] They share a large amino-terminal domains with putative helicase/primase elements features that are known only in the thermophilic viruses and Aquificae. A horizont transfer seems the most likely explanation for these findings.
The telomerase (tert) is a large protein (2518 codons) and has a predicted molecular weight of ~280 kiloDaltons.[291] It has the usual telomerase specific motifs within the N-terminal half of the protein (GQ/N, CP, QFP and T) and reverse transcriptase (RT) specific motifs in the C-terminal half. The N-terminal half is required for efficient binding of the RNA template, defining the 5′ RNA template boundary, multimerization and interactions with associated proteins. The RT domain is essential for the catalytic activity. The protein contains several nuclear localization signals and is found in the nucleolus.
A putative tyrosine site specific recombinase has been isolated.[517] The N-terminus has the typical alpha helical bundle and potentially a mixed alpha-beta domain resembling that of λ-Int. The C-terminal domain has the putative tyrosine recombinase conserved active site residues Lysine-Histadine-Lysine-(Histadine/Tryptophan)-Tyrosine. The gene is expressed differentially during the erythrocytic stages being maximal in the schizont stage. The open reading frame encodes a ∼57 kiloDalton protein. Knockout mutants are viable and appear normal. DNA binding studies suggest a number of targets include the subtelomeric regions.
A number of mini chromosome maintenance proteins are present in the genome.[518] These are large proteins and members of the AAA ATPase family with a conserved region of ~200 amino acids responsible for nucleotide binding. They are responsible for unwinding DNA at the replication forks and are involved in other chromosome transactions such as transcription, chromatin remodeling and genome stability.
A SIP2 gene, a member of the ApiAP2 family of putative transcription factors, has been cloned.[519] It appears to be involved in maintenance of the chromosome ends rather than in the regulation of particular genes.
A novel DNA/RNA binding protein PfAlba has been described.[520] This protein is related to the archaeal protein Alba (Acetylation lowers binding affinity). There are at least four paralogs of the PfAlba gene and these proteins form a complex with the P. falciparum specific TARE6 (Telomere-Associated Repetitive Elements 6) subtelomeric regions. Also associated with the TARE6 regions are PfSir2 a histone deacetylase. In the early blood stages the PfAlba proteins are enriched at the nuclear periphery and associate with the PfSir2 proteins. When the parasite switches from trophozoite to the schizont stage the PfAlba proteins move to the cytoplasm. These proteins will also bind single stranded RNA but the reason for this binding is not known.
The single stranded DNA binding protein (SSB) plays an important role in all known organisms. A SSB protein is encoded in the genome and localises to the apicoplast.[521] It forms a homo-tetramer alone and when bound to single stranded DNA. The protein binds 52-65 nucleotides/tetramer.[522] While similar in its overall structure to that of the SSB of E. coli it differs at the carboxy terminal region. Although it binds single stranded DNA in a similar fashion to the SSB of E. coli it does so with the opposite polarity. There are a number of other functional differences between this protein and that of E. coli. The basis for these differences has yet to be determined.
A protein - RPA1L - is the homologue of the bacterial single stranded binding protein (SSB) and acts in initiating homologous pairing and strand exchange activity.[523] It is negatively regulated in a dose dependent manner by RPA1S.
The eukaryotic homologue of the bacterial RecA protein is Rad51. The Plasmodium falciparum Rad51 protein exhibits ATPase activity and promotes DNA strand exchange.[523] This protein interacts with Rad54 and replication protein A.
SET is a conserved nuclear protein involved in chromatin dynamics.[524] In P falciparum it is expressed in both asexual and sexual blood stages but strongly accumulates in male gametocytes. In P falciparum there are two distinct promoters upstream. One is active in all blood stages while the other is active only in gametocytes and in a fraction of schizonts possibly committed to sexual differentiation. In ookinetes both promoters exhibit a basal activity, while in the oocysts the gametocyte specific promoter is silent and the reporter gene is only transcribed from the constitutive promoter.
Plasmodium appears to lack both DNA methylation and the RNA interference machinery.[94][525]
DDX6/DOZI (development of zygote inhibited) is a member of the DEAD box family and is involved in the sexual development of the protozoan parasite.[526] The gene is known as PfDZ50 in P. falciparum. It binds DNA and RNA and has nucleic acid dependent ATPase and RNA unwinding activities. It interacts with eIF4E mainly through domain 1 and inhibits translation. It is localized mainly in the granular bodies found throughout the cytoplasm during the asexual intraerythrocytic developmental stages.
A novel transcription factor - PREBP - has been described.[527] This protein has 4 KH homology domains which are found in RNA-binding or single-stranded DNA-binding proteins. PREBP is well conserved in Plasmodium species and partially conserved in phylum Apicomplexa. It acts on the pf1-cys-prx, a gene expressed in the trophozoite/schizont stages.
- DNA repair proteins
Several nucleic acid repair pathways are known. These include the nucleotide excision repair, the mismatch repair, the base excision repair, the double strand break repair and the cross link repair pathways. DNA replication errors - base substitution mismatches and insertion-deletion loops - are primarily corrected by the mismatch repair system.
The MutL homolog (MLH) - part of the DNA mismatch repair system - has been cloned.[528] MLH possess ATPase and endonuclease activities. Its expression is maximal in the schizont stage.
Polynucleotide kinase/phosphatase (PNKP) is a bifunctional enzyme that can phosphorylate the 5'-OH termini and dephosphorylate the 3'-phosphate termini of DNA. It is a DNA repair enzyme involved in the processing of strand break termini, which permits subsequent repair proteins to replace missing nucleotides and rejoin broken strands. A P. falciparum gene encoding a protein with 24% homology to human PNKP has been cloned.[529] This enzyme dephosphorylates single-stranded substrates or double-stranded substrates with a short 3'-single-stranded overhang, but not double-stranded substrates that mimicked single-strand breaks.
- RNA binding proteins
The messenger RNA capping system appears to be more similar to that of fungi than to vertebrates.[530] It encodes RNA guanylyltransferase (Pgt1) and RNA triphosphatase (Prt1) enzymes. The triphosphatase enzyme is a member of the fungal/viral family of metal dependent phosphohydrolases. These are structurally and mechanistically unrelated to the cysteine phosphatase type RNA triphosphatases found in metazoans and plants.
A protein (PfSR1) involved in alternative splicing has been described.[531] It appears that regulation of this gene is essential for the parasite's normal physiology.
A number of the DExD/DExH-box containing pre-mRNA processing proteins (Prps) - PfPrp2p, PfPrp5p, PfPrp16p, PfPrp22p, PfPrp28p, PfPrp43p and PfBrr2p - are present in the genome.[532] PfPrp16p a helicase and a member of DEAH-box protein family with nine collinear sequence motifs has been cloned. It binds to RNA, hydrolyses ATP and appears to be involved in splicing.
The RNA polymerase II has an unual expansion in the C terminal domain.[533] The primate infecting species posses a larger number of repeats that are found in either the bird or rodent species.
A cyclin (Pfcyc-1) has been cloned and its structure has been solved.[534] The protein has a typical cyclin box which consists of two tandemly repeating five-helix bundles separated by a linker hinge peptide.
The origin recognition complex subunit 1 (Orc1) associates with fSir2 in the sliencing complex.[535]
At least seven proteins are known to be involved in the splicing process.[536] They are expressed at asexual blood stages of the parasite, show nucleo-cytoplasmic localization and form a ring like heptameric complex like other eukaryotes. The interaction of PfSMN (survival of motor neuron, Tudor domain containing protein) or PfTu-TSN (tudor domain of Tudor Staphylococcal nuclease) with the PfSmD1 protein is methylation dependent. The arginine methylase PfPRMT5 appear to interact with PfSmD1 suggesting a role for arginine methylation in the assembly of the spliceosome complex.
Helicases
An unusual helicase - a homologue of Dbp5 and DDX19 from yeast and human respectively - has been cloned.[537] It possesses DNA and RNA unwinding, nucleic acid dependent ATPase and RNA binding activities. A Q motif is required for its activity.
Another DEAD box helicase - a homolog of Has1p from yeast which has DNA and RNA unwinding, nucleic acid dependent ATPase and RNA binding activities - has also been cloned.[538]
The DNA helicase II (uvrD) is a superfamily 1A helicase which plays an essential role in the mismatch repair pathway. Homologs of UvrD include the proteins PcrA and Rep. These proteins have a two domain (1 and 2) structure with each domain made of two sub-domains (1A, 1B, 2A and 2B) and a C-terminal extension. They are DNA-dependent ATPases with 3′ to 5′ helicase activity. The helicase activity is located in the N terminal domain.
The UvrD protein of P. falciparum has been cloned.[539] This gene (PFE0705c) is located on chromosome 5 and contains no introns. It is 4326 bases in length, encodes a protein of 1441 amino acids and has a predicted molecular weight of ~170 kiloDaltons. The two domains and their subdomains are present: The 1A domain is from amino acid 1–722; the 1B domain is from amino acid 150–464; the 2A domain is from amino acid 723–1441; and the 2B domain is from amino acid 896–1359. There is no C-terminal extension. The ATPase and helicase activity are confined to domain 1A and 1B (the N-terminal and first half of the C terminal). It is expressed in the schizont stages of intraerythrocytic development and it colocalizes with PfMLH, a protein involved in mismatch repair. Both PfDH60 - another helicase - and PfMLH are also expressed in schizont stages.
A helicase - PfH45 - of 398 amino acid residues (molecular weight 45 kiloDaltons) is a unique bipolar helicase with both the 3' to 5' and 5' to 3' directional helicase activities.[540] It is expressed in all the intraerythrocytic developmental stages and has a role in translation.
A 3'-5' DNA helicase has been identified in the parasite.[541] The apparent molecular weight is 90 kiloDaltons. Its activity is dependent on the presence of magnesium and ATP.
A homolog of UAP56 (U2AF65 associated protein) - a member of the DEAD box helicase family - has been cloned.[542] This homolog - PfU52 - contains the RNA dependent ATPase, RNA helicase and RNA binding activities. This protein is expressed in all the intraerythrocytic developmental stages of the parasite. Residues glycine 181, isoleucine 182 and arginine 206 are involved in RNA binding and this binding activity is required for its enzymatic activities.
The RuvB proteins belong to AAA+ family of enzymes which are involved in diverse cellular activities. There are at least 3 copies of this protein in the genome.[539] The PfRuvB1 protein has considerable homology with human as well as yeast RuvB1 and contains both Walker motif A and Walker motif B.[543] It is an ATPase and this activity increased significantly in the presence of single stranded DNA. It also has DNA helicase activity and translocates preferentially in 5' to 3' direction. It is constitutively expressed during all the stages of intraerythrocytic cycle and localizes mainly to the nucleus.
PfRuvB2 similarly has both ATPase and weak DNA helicase activities.[544] It is expressed in all the asexual intraerythrocytic developmental stages and localizes mainly in the nucleus during merozoite, ring and trophozoite stages while during schizont stage it relocalizes partially in the nucleus and partially towards cytoplasm. It interacts with PfRuvB3 and not with PfRuvB1.
RuvB3 possesses the Walker motif A, Walker motif B, sensor I and sensor II conserved motifs similar to yeast and human RuvB like proteins. It has single stranded DNA dependent ATPase activity. The protein is mainly expressed during intraerythrocytic schizont stages and localizes to the nuclear region. In the merozoite the protein relocalizes to the sub nuclear region. The PfRuvB2/PfRuvB3 complex preferentially translocates and unwinds DNA in the 5'-3' direction.
Histones and their modifying enzymes
Histones are essential for the correct packing and function of DNA in eukaryotes. Variants of these proteins are known to occur throughout eukaryotes and these are thought to play a role in epigenetic control. Although variants of H2A and H3 are common, variants of H2B and H4 are much less so. H2B.Z is an apicomplexan specific H2B variant.[545] H2B.Z localises to euchromatic intergenic regions throughout intraerythrocytic development and with H2A.Z may form nucleosomes. These H2A.Z and H2B.Z nucleosomes are more common in promoters over 3' intergenic regions and their occurrence is related to the promotor's strength. The presence of H2B.Z is correlated with the base composition of the underlying DNA.
Other variants of histones known to occur in this genome are H2Bv, H3.3 and CenH3.[546] Covalent modification of the histones is well known with at least 44 different types having been described.[547] The deacetylation and subsequent tri-methylation of lysine 9 on histone H3 (H3K9me3) as well as the recruitment of heterochromatin protein 1 are found in heterochromatic islands and are involved in the regulation of the var genes.[548] Other modifications that have been described include H3K4me3, H3K9ac, H3K14ac and H4ac.[547] H2A.Z is consistently found at both 5′ and 3′ ends of the euchromatic genes. The histone acetyl transferases (Gcn5 and Esa1) that can act on H2A.Z are present in the genome.[549]
Posttranslational covalent modifications of the histones (acetylation, methylation and phosphorylation) are known to occur. How these influence gene transcription is poorly understood. The 14-3-3 protein appears to act as a phosphorlyated histone 3 reader.[421]
A histone deacetylase (HDAC1) has been cloned.[550] The protein has 449 amino acid residues and localises to the nucleus. Its molecular weight is 50 kiloDaltons and it is predominantly expressed in mature asexual blood stages and in gametocytes.
Sir2A is a member of the sirtuin family of nicotinamide adenine dinucleotide dependent deacetylases. In P. falciparum it has been has been shown to regulate the expression of surface antigens to evade the detection by host immune surveillance.[551] While it is a poor deacetylator of histones it also catalyzes the hydrolysis of medium and long chain fatty acyl groups from lysine residues. Proteins are present in P. falciparum with these modifications and these can be removed by can be removed by PfSir2A in vitro. This suggests that this may be its role rather than the deacetylation of histones.
During the life cycle the telomeres and telomere associated repeat elements are transcribed as long non coding RNAs.[552] They are transcribed by RNA polymerase II as single-stranded molecules. In the ring stage, these transcripts are located in a single perinuclear compartment that does not co-localize with any known nuclear subcompartment. During the schizont stage they are found at several nuclear foci. At least some of these can form stable and repetitive hairpin structure that is able to bind histones. Their function requires further elucidation.
Reversible histone modifications can cause changes in gene expression and alter the phenotype.[376]
Gene regulation
A number of novel DNA binding sites have been identified along the genome.[553] Their function - if any - remains to be determined.
Apetala 2 (AP2) family proteins are transcription factors that have DNA-binding domains of ~60 amino acids called AP2 domains. 27 AP2-family genes have been identified in the Plasmodium falciparum genome.[554] One of these proteins appears to play a critical role in the liver stage development of the parasite.[555]
The transcription factor NF-YB is localised in the nucleus during the erythrocytic stages of the life cycle.[556] Melatonin and cyclic adenosine monophosphate modulate the expression of NF-YB. NF-YB is also more ubiquitinated in the presence of melatonin.
The protein PfMyb1 is a transcription factor belonging to the tryptophan cluster family.[557] Inhibition of this gene reduces growth by ~40% with the mortality being concentrated at the trophozoite-schizont interface.
SAP1 has been shown to be involved in the post transcriptional control of liver stage genes.[558]
CCR4-associated factor 1 is involved in the regulation more than 1000 genes during malaria parasite's intraerythrocytic stages.[559] Mutations in this gene result in mistimed expression, aberrant accumulation and localization of proteins involved in parasite egress and invasion of new host cells. This leads to the premature release of predominantly half-finished merozoites in turn drastically reducing the intraerythrocytic growth rate of the parasite.
Mutation rates
The overall mutation rate of the genome is 1.0-9.7 x 10-9 mutations per base pair per generation.[560] The rate in genes involved in immune evasion is higher - 9.5 x 10-6 structural variants per base pair per generation.
Protein metabolism
It has been hypothesized that the parasite obtains all, or nearly all, of its amino acids by salvaging from the host or through the degradation of hemoglobin. This is supported by the fact that genomic analysis has found no enzymes necessary for amino acid biosynthesis, except for glycine-serine, cysteine-alanine, aspartate-asparagine, proline-ornithine, and glutamine-glutamate interconversions.[3]
Deoxyhypusine hydroxylase catalyses the final stage of the synthesis of the amino acid hypusine. In eukaryotes this amino acid is only found in eukaryotic initiation factor 5A. This gene has been identified in the genome of P. falciparum, P. knowlesi, P. vivax and P. yoelii.[561]
Deoxyhypusine synthase catalyzes the first step in hypusine biosynthesis of eukaryotic initiation factor 5A (EIF5A). Inhibitors of this enzyme may be of use therapeutically.[562]
Much of the digestion of haemoglobin occurs within the digestive vacuole. Multiple enzymes are involved in this process including four distinct plasmepsins (aspartic acid proteases).
Protein translation
There are two translation elongation factor G proteins encoded in the genome.[563] One is located in the mitochondrion and the second in the plastid. Both appear to be inhibitable with fusidic acid
Two Ribosome Recycling Factors (RRF1 and RR2) are present in the genome.[264] Both proteins are targeted to both the apicoplast and the mitochondrion. RRF2 is also present in the cytoplasm. Unusually it forms dimers. RRF1 has a 108 amino acid insert compared with that of other organisms. The function of this insert - if any - is currently unknown.
The eukaryotic translation initiation factor 2α has a regulatory serine at position 51. This can be phosphorylated by several kinases. Three are known in P falciparum: IK1, IK2 and PK4.[564] IK1 regulates stress response to amino acid starvation; IK2 inhibits development of malaria sporozoites present in the mosquito salivary glands; and PK4 is essential for the completion of the parasite's erythrocytic cycle.
Unlike other organisms, Plasmodium codon bias is not correlated to tRNA gene copy number.[565]
Aminoacyl tRNA synthetases are required for protein synthesis. Alanine tRNA synthetase, glycine tRNA synthetase and threonine tRNA synthetase are dually localised to the cytosol and the apicoplast.[566] These enzymes do not appear to be present in the mitochondrion.
Tyrosyl tRNA synthetase is secreted by the parasite into the cytoplasm of the infected erythrocyte.[567] On lysis of the erythrocyte it is released into the blood stream where it is pro inflammatory. It is specifically bound by and taken up by host macrophages and leads to enhanced secretion of the cytokines tumor necrosis factor-alpha and interleukin 6. This interaction also increases the adherence linked host endothelial receptors ICAM-1 and VCAM-1.
The cytoplasmic lysyl-tRNA synthetase is dimeric unlike the human version which may be dimeric or tetrameric.[568] It is capable of synthesizing the signalling molecule diadenosine tetraphosphate using ATP as a substrate.
The tryptophanyl-tRNA synthetase appears to be localised to the cytoplasm.[568] This enzyme has an unusual N-terminal extension that appears to be essential for its activity.
Post translational modifications
Palmitoylation - the reversible addition of a lipid moiety to a cysteine residue - appears to be common in this parasite.[569] Its role in its biology is not yet understood.
An N-myristoyltransferase is present in the genome.[570] It is involved in protein trafficking.
Lysine acetylation appears to be common in this organism.[61] Lysine-acetylated proteins are present in the nucleus, cytoplasm, mitochondrion and apicoplast. The acetyltransferase PfMYST appears to be involved in this process. The effects of these modifications are not yet understood.
Hemoglobin metabolism
During the erythrocytic stage of the parasite's life cycle, it uses intracellular hemoglobin as a food source. The protein is broken down into peptides, and the heme group is released and detoxified by biocrystallization in the form of hemozoin.[571]
Heme biosynthesis by the parasite has been reported.[572] Mutations have been induced in both the first and last enzymes (δ-aminolevulinate synthase and ferrochelatase) of this pathway.[573] Although these mutations do not appear to have an effect on the growth in blood cells, growth in both the mosquito and in the liver requires an intact pathway. The muatated strains form decreased numbers of oocyts and fail to form sporozoites. These strains fail to be infective to hosts. The parasites incorporate both host haemoglobin heme and parasite synthesized heme into haemozoin and into the mitochondrial cytochromes.
Haemozoin
Haemozoin (malarial pigment) is a ferriprotoporphyrin IX crystal produced by Plasmodium parasites after haemoglobin catabolism. It is an insoluble phase of iron (III) protoporphyrin-IX. The structure of haemozoin was solved in 2000 by powder X-ray diffraction.[574] It is a crystalline structure composed of heme units interlinked to form cyclic dimers via reciprocal iron-oxygen (propionate) bonds.[286] Its nucleation occurs at the inner membrane of the digestive vacuole, with crystallization occurring in the aqueous rather than lipid phase.[286] Acylglycerol lipids are involved in the nucleation process.
Ferriprotoporphyrin IX (haematin) competes with NADH for the active site of the enzyme lactate dehydrogenase.[575] This competition may be fatal to the parasite. To detoxify the ferriprotoporphyrin IX the parasite polymerizes haematin to haemozoin.
The conversion of haemoglobin to haemozoin is mediated by a ~200 kiloDalton protein complex within the digestive vacuole.[576] This complex contains a number of proteins including falcipain 2/2', plasmepsin II, plasmepsin IV, histo-aspartic protease and heme detoxification protein. The proteins spontaneously associate with each other and can convert haemoglobin to haemozoin. The heme detoxification protein has two heme-binding sites.[577] The drugs chloroquine and artemisinin both act during the heme polymerization step. Chloroquine also acts at the haemoglobin degradation step.
Although the parasite has a heme oxygenase like protein which binds bound heme and protoporphyrin IX with modest affinity, this protein does not catalyze heme degradation in vivo.[578] It lacks a critical heme-coordinating histadine residue and this probably accounts for its lack of heme oxygenase activity. The function of this protein remains unknown.
- Drugs interfering with haemozoin formation
Artemisinin an anti malarial agent appears to require haemoglobin digestion for its activity.[579] Inhibition of hemoglobinase activity with cysteine protease inhibitors, knockout of falcipain 2 or deprivation of host cell lysate reduce the activity of the drug against the parasite.
Chloroquine causes a dose dependent decrease in haemozoin by disrupting hemozoin crystal growth.[580] This results in mosaic boundaries in the crystals formed in the parasite. This disruption causes a redistribution of heme from the parasite digestive vacuole to the its cytoplasm.
Other drugs that inhibit haemozoin formation include amodiaquine, artesunate, lumefantrine, mefloquine and quinine.[580] Neither pyrimethamine nor sulfadoxine affect haemazoin formation.
- Effects on the host
Haemozoin has multiple effects on the host's immune system including increasing the level of matrix metalloproteinase-9.[581] Its presence within the lung is correlated with lung inflammation and it may be important in the pathogensis of acute respiratory distress syndrome in malaria.[582]
Haemozoin has been shown to promote inflammation mediated lysozyme release from human monocytes through p38 mitogen activated protein kinase and nuclear factor κB dependent mechanisms.[583] This process may also involve 15-hydroxyeicosatetraenoic acid.
Both matrix metalloproteinase 9 and its endogenous inhibitor - tissue inhibitor of metalloproteinase 1 - are induced in macrophages by haemozoin.[583] Both p38 MAPK and NF-κB appear to be involved in this induction pathway.
Haemazoin is recognised by a number of immune receptors including the Toll-like receptors.[584] Phagolysosomal formation within the macrophages after haemozoin ingestion is normal but the haemozoin remains stored inside these cells for months or even longer without any detectable degradation.
Haemozoin inhibts erythropoetin induced proliferation of erythroid precursors.[585] The mechanism is unknown but this probably contributes to the anemia associated with malaria.
Carbohydrate metabolism
During erythrocytic stages, the parasite produces its energy mainly through anaerobic glycolysis, with pyruvate being converted into lactate.[3]
Genes encoding for the TCA cycle enzymes are present in the genome, but it is unclear whether the TCA cycle is used for oxidation of glycolytic products to be used for energy production, or for metabolite intermediate biosynthesis.[3] It has been hypothesized that the main function of the TCA cycle in P. falciparum is for production of succinyl-CoA, to be used in heme biosynthesis.[3]
Genes for nearly all of the pentose phosphate pathway enzymes have been identified from the genome sequence.
Glycerol is a major glucose metabolite.[586]
Pools of several sugars (UDP-glucose, UDP-galactose, UDP-N-acetylglucosamine, GDP-mannose and GDP-fucose) are present in the cytoplasm of the blood stages.[587] The enzymes GDP-mannose 4,6-dehydratase and GDP-L-fucose synthase are present in the genome.
Both the asexual and gametocyte stages of the life cycle utilise the Kreb's cycle to generate adenosine triphosphate.[588] Glutamine is the preferred substrate over glucose in the asexual stages while in the gametocytes, glucose is preferred. Inhibition of this cycle in the asexual stages has little effect while inhibition in the gametocytes is lethal.
The enzyme hexokinase limits the rate of entry of glucose into glycolysis.[589]
Mutations in the glycerol kinase gene limit the production of glycerol-3-phosphate which is used in phosphlipid biosynthesis.[590] Exogenous glycerol may be used as an alternative source.
Lipid metabolism
Phosphatidylcholine is a major and essential membrane phospholipid in the parasite. Its synthesis occurs via the CDP-choline and the serine decarboxylase phosphoethanolamine methylation pathways. The substrates of these pathways are the host's choline, serine and fatty acids. Both pathways share the final two steps catalyzed by two essential enzymes CTP:phosphocholine cytidylyltransferase and choline-phosphate transferase. Mutations in phosphoethanolamine N-methyltransferase are associated with severe alterations in gametocyte development, are incapable of producing mature-stage gametocytes and are not transmittable to mosquitoes.[591]
The major source of phosphatidylcholine is the CDP-choline Kennedy pathway.[592] Both phosphoethanolamine-Nmethyltransferase and phosphatidylethanolamine-N-methyltransferase are important in its synthesis. Most of the serine derived phosphatidylethanolamine is formed via serine decarboxylation, whereas the majority of phosphatidylserine is formed by base exchange reactions.
The other major component of the parasite's membranes is the phospholipid phosphatidylethanolamine.[593] This phophlipid is not found in the erythrocyte. Phosphatidylethanolamine is synthesised de novo by the CDP-ethanolamine dependent Kennedy pathway. The rate limiting step in this pathway is the step involving CTP:phosphoethanolamine cytidylyltransferase. This enzyme is composed of two cytidylyltransferase domains separated by a linker region. Only the N-terminal domain appears to be enzymatically active. The enzyme appears to function as a dimer and to obey Michaelis-Menten kinetics.
A phosphatidylserine decarboxylase has been cloned from the parasite Plasmodium knowlesi.[594] It seems highly probably this enzyme is also found in P. falciparum.
β-hydroxyacyl-acyl carrier protein dehydratase catalyzes the third and important reaction of the fatty acid elongation cycle. The P. falciparum gene has been cloned and the crystal structure of the enzyme solved.[595]
α-lipoic acid (6,8-thioctic acid: LA) is a vital co-factor of α-ketoacid dehydrogenase complexes and the glycine cleavage system.[596] It is essential for the erythrocytic and liver stages of Plasmodium and is the co-factor of the acetyltransferase subunit of pyruvate dehydrogenase located in the apicoplast.
LA biosynthesis, comprising octanoyl-acyl carrier protein (ACP): protein N-octanoyltransferase and lipoate synthase is exclusively found in the apicoplast where it generates LA de novo from octanoyl-ACP, provided by the type II fatty acid biosynthesis pathway which is also present in this organelle.[596]
Other members of the fatty acid synthesis type II pathway present in the genome are PfFabI, PfFabG and PfFabZ.[597]
Two lipoic acid protein ligases (LplA1 and LplA2) are present in the genome.[596] LplA1 is confined to the mitochondrion while LplA2 is found in both the mitochondrion and the apicoplast. LplA1 exclusively uses salvaged LA and lipoylates α-ketoglutarate dehydrogenase, branched chain α-keto acid dehydrogenase and the H-protein of the glycine cleavage system. LplA2 cannot compensate for the loss of LplA1 function during blood stage development.
The parasite syntheses 2-C-methyl-d-erythritol-4-phosphate. Other enzymes in the this pathway include 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase and 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate synthase and 1-deoxy-D-xylulose 5-phosphate synthase (the first enzyme in the pathway).[67] This pathway is responsible for the production of the essential isoprenoid precursors, isopentenyl diphosphate and dimethylallyl diphosphate.
The enzyme 1-deoxy-D-xylulose-5-phosphate reductoisomerase can be inhibited by the antibiotic fosmidomycin which has been shown to potentially useful as an antimalarial.[598]
Isoprenoid biosynthesis may be inhibited by fosmidomycin which in turn reduces protein prenylation. One of the proteins that is normally prenylated is Rab5 - a protein involved in vesicle transport. On inhibition by fosmidomycin the Rab5 proteins mislocalize and cause marked defects in food vacuolar morphology and integrity.[599]
HMB-PP synthase (IspG), an iron-sulphur (4Fe4S) protein involved in isoprenoid biosynthesis, has two domains - a TIM barrel and a 4Fe4S domain - in bacteria. In plants and malaria parasites, there is an additional large insert domain.[600] This is a second TIM barrel that interacts with the other TIM barrel.
Rather than using sphingomyelin as the primary complex sphingolipid, yeast, plants and some protozoa utilise an evolutionarily related inositol phosphorylceramide synthase to synthesize inositol phosphorylceramide. In the P. falciparum genome there is a single copy of a putative sphingomyelin synthase.[601] This gene's homolog in Toxoplasma gondii is the functional orthologue of the yeast's inositol phosphorylceramide synthase.
The parasite's requirement for acetyl-CoA are supplied by several pathways including acetyl-CoA synthetase and a pyruvate dehydrogenase like enzyme.[258] The nature of the pyruvate dehydrogenase like enzyme is not quite clear but it seems likely that the branched chain ketoacid dehydrogenase complex is performs this function. This latter pathway contributes glucose derived acetyl-CoA to the tricarboxylic acid cycle in a stage independent process whereas anapleurotic carbon enters the this cycle via a stage dependent phosphoenolpyruvate carboxylase / phosphoenolpyruvate carboxy-kinase process that decreases as the parasite matures.
- Membrane biogenesis
Membrane biogenesis in this organism involves the enzyme phosphoethanolamine methyltransferase which catalyses the methylation of phosphoethanolamine to phosphocholine. This pathway is found in plants and nematodes but not in humans. The enzymes in P. falciparum is a multi-functional unlike that of plants and nematodes.[602] The enzyme from P. falciparum has been cloned and its structure solved.
Four putative autophagy genes - those involved in ATG8 lipidation - are present in the genome.[603] Atg8 lipidation requires Atg7 (an E1-type ligase), Atg3 (an E2-type ligase) and Atg4 (a cysteine protease). PfAtg7 is the activating enzyme of PfAtg8. PfATg7 appears to be essential for normal growth.
Trace metals
Zinc is essential to the parasite and it actively accumulates it during growth.[604] During the life cycle zinc pools are formed in both the cytosol and the mitochondria.[605]
A copper channel has been identified in the genome.[606] Removal of copper from the growth medium is associated with early developmental arrest.
Vitamin A
The parasite takes up vitamin A from its host and uses it in own metabolism. Children with malaria have lower vitamin A levels than normal.[607]
Miscellaneous proteins
Polyamine biosynthesis in these parasites is controlled by a unique bifunctional S-adenosylmethionine decarboxylase/ornithine decarboxylase (PfAdoMetDC/ODC).[608] On the secondary structure level PfAdoMetDC is similar to that of the human protein. This bifunctional enzyme ensure coordination decarboxylated AdoMet and putrescine for the subsequent synthesis of spermidine.
The first two reactions of the pentose phosphate pathway in P. falciparum are catalysed by a single bifunctional enzyme - glucose 6-phosphate dehydrogenase 6-phosphogluconolactonase.[609] This is distinct from the case in humans where the enzymes are separate. In animals this pathway is usually found in the cytosol while in plants it is found in the plastids. The location of this reaction is not currently known in P. falciparum.
Fusions between these two enzymes (glucose 6-phosphate dehydrogenase and 6-phosphogluconolactonase) have also been reported in chordates.[610] The chordate fusion differs in its orientation to that in Plasmodium (in Plasmodium the 6-phosphogluconolactone is found at the N-terminus of the glucose 6-phosphate dehydrogenase protein) indicating that at least two separate fusion events have occurred. The metazoan fusion appears to have occurred near the bases of the metazoan and apicomplexan lineages. This fusion event was not found in any of the three sequenced Cryptosporidium genomes. It was not found in Perkinsus marinus or in either of the ciliate (Paramecium tetraurelia and Tetrahymena thermophila) genomes. More data will be needed to estimate the timing of this fusion event.
Only one of the two metazoan paralogs of glucose 6-phosphate dehydrogenase is fused, indicating that the fusion occurred after a duplication event. This duplication event occurred in an ancestor of the choanoflagellates and metazoans. Another fusion event between these enzymes occurred in an ancestor of the protozoan parasites Trichomonas and Giardia lamblia. In Giardia, the proteins are fused in opposite orientations. A third fusion event occurred between glucose 6-phosphate dehydrogenase with phosphogluconate dehydrogenase in a diatom species (Phaeodactylum tricornutum).
The mechanism of action of the triose phosphate isomerase enzyme has been investigated in some detail.[611] The conserved glutamic acid residue at position 97 is involved in the catalytic proton transfer. Modification of this residue may reduce the rate of catalysis by 9000 fold.
The shikimate pathways is functional in P. falciparum and vitamin E biosynthesis also occurs.[612]
The parasite actively synthesises pyridoxal-phosphate (vitamin B6). The 2-C-methyl-d-erythritol-4-phosphate pathway is involved in its synthesis. This process involves two sets of reactions: condensation of ribulose 5-phosphate, glyceraldehyde-3-phosphate and ammonia produced from glutamine. These actions are carried out by separate subunits. The synthase domain is known as Pdx1 and the glutaminase domain as Pdx2. In P. falciparum the core Pdx1 is a dodecamer and forms the core of the enzyme. There are up to 12 Pdx2 subunits surrounding the Pdx1 subunit.[613] The majority of the synthesis is carried out by Pdx1. The pentose substrate is covalently attached through its C1 and forms a Schiff base with the Lysine 84 residue. The ammonia transfer between Pdx2 glutaminase and Pdx1 active sites is regulated by a transient tunnel.
Chorismate synthase (CS) catalyses the seventh and final step of the shikimate pathway. P. falciparum chorismate synthase (PfCS) is unique in terms of enzymatic behavior, cellular localization and in having two additional amino acid inserts compared to any other CS.[614]
There are several versions of the enzyme glutamate dehydrogenase (GDH) encoded in the genome. Of these, GDH1 and 3 appear to localise in the cytoplasm and GDH2 to the apicoplast.[615]
Within the genome, there are two adenylyl cyclases - ACα and ACβ.[616] ACα contains six predicted transmembrane domains and a single carboxy-terminal catalytic domain homologous to sAC-like ACs. It is a predicted bifunctional protein comprising both a potassium channel and an AC that is conserved in the alveolates. It is expressed in the gametocytes. ACβ has no predicted transmembrane regions and possesses two AC catalytic domains. It has a marked pH dependence and is required for the erythrocytic stages.
Asymmetrical diadenosine 5',5″-P1,P4-tetraphosphate hydrolase (EC 3.6.1.17) catalyses the conversion of diadenosine 5',5″-P1,P4-tetraphosphate (Ap4A) to ATP and AMP and diadenosine 5',5″-P1,P5-pentaphosphate (Ap5A) to ATP and ADP. This enzyme from the parasite has been cloned and expressed.[617]
A glycerophosphodiesterase has been cloned.[618] This enzyme is found in the parasitophorous vacuole, digestive vacuole and cytosol. It appears to be an essential gene but its specific function is currently unclear.
The translationally controlled tumor protein appears to bind artemisinin.[619] This may contribute to its anti malarial action.
A bifunctional farnesyl diphosphate/geranylgeranyl diphosphate synthase has been cloned.[620] It is encoded by the gene PF3D7_1128400.
Thiamine appears to be essential to the parasite's metabolism.[621] Thiamine is converted to its active form - thiamine pyrophosphate - by thiamine pyrophosphokinase. The enzymes oxoglutarate dehydrogenase and pyruvate dehydrogenase are both dependent on thiamine pyrophosphate.
A thymidylate kinase - an enzyme that catalyzes phosphorylation of thymidine monophosphate to thymidine diphosphate - is present in the genome.[622]
Human immune system evasion
var family
The var genes encode for the P. falciparum erythrocyte membrane protein 1 (PfEMP1) proteins. The genes are found in the subtelomeric regions of the chromosomes. There exist an estimated 59 var genes within the genome.[3] The proteins encoded by the var genes are ultimately transported to the erythrocyte membrane and cause the infected erythrocytes to adhere to host endothelial receptors. Due to transcriptional switching between var genes, antigenic variation occurs which enables immune evasion by the parasite.
The hypervariable var gene repertoire is to a large extent generated by frequent meiotic ectopic recombination in the mosquito gut. Mitotic recombination may also occur.[623]
- Gene structure
The var genes are classified into a number of subfamilies (A, B and C) that possess distinctive upstream and downstream flanking regions. A number of intermediate types - B/A and B/C - have also been described. The classification system is based primarily on the orientation of the genes: subtelomeric A and B family genes are oriented tail to tail (3′ to 3′) while central C family genes are oriented head to tail in a tandem repeat manner. The two intermediate groups - B/A and B/C- have chromosomal positions or domain composition that differ from those that would be expected from their orientation.
From N- to C-terminal the genes are organised in the following fashion: an N-terminal segment (NTS), Duffy binding-like (DBL) domains, Cysteine rich inter-domain regions (CIDR), C2 domains, one transmembrane region (TM) and the acidic terminal segment (ATS).
The extracellular region of PfEMP1 comprises multiple adhesion domains called Duffy Binding Like (DBL) and Cysteine Rich Interdomain Region (CIDR). The number of DBL domains in each var gene varies between 2 and 9: the usual number is ~6. The DBL domains have been classified into different major classes - α, β, γ, δ, ζ and ε - and a number of sub-classes based on sequence criteria. Within the DBL are seven regions of considerable variability known as variable blocks (VB1-6). In all 147 subtypes have been recognised. In addition to these 21 conserved tandem runs of specific domains (domain cassettes) have also been identified.[624] The DBL domains have been also characterized by definition of 10 semi-conserved homology blocks (HBa-j) interspersed by hypervariable regions and by the definition of three structural subdomains (S1–3). Some biological correlations with the DBL domains have been described: DBLα has been associated with binding to heparin sulfate, blood group A antigen and complement receptor 1; DBLβ domains have affinity for ICAM-1; and DBLδ adheres to platelet-endothelial cell adhesion molecule 1 (PECAM-1).[625]
The CIDR domains have been divided into three classes - α, β, and γ - and have three regions: the minimal CD36 binding region (M2) which is flanked flanked by the less conserved M1 and M3 regions.[626] Several CIDRα class domains mediate binding to the human CD36 receptor but this binding only occurs in the B and C var families.[627] The CIDRα domains also bind immunoglobulin M and PECAM-1.[625]
An association between the length of the gene and its sequence variation has been reported.[628] Shorter genes tend to be more variable and long genes to be more conserved. This probably is the result of a trade off between optimizing within host fitness and minimizing between host immune selection pressure.
- Evolution
These genes appear to have evolved before the separation Plasmodium falciparum and Plasmodium reichenowi.[629]
- Gene expression
The var genes undergo antigenic switching - only a single gene is expressed at a given time and the expression varies during the course of infection. This variation begins when the merozoites leave the liver and the mechanisms driving this process are not presently understood. A method of erasing the epigenetic memory which appears to be involved in this process has been described.[630] An analysis of the results showed that a subtype of var genes - the upsA vars - which are rarely expressed in culture systems but appear to be important in clinical infections, are activated early in the switching process. The switching rate appears to be a function of the gene structure rather than its chromosomal position or promoter.
The protein PfSir2 associates with promoter regions of silenced genes involved in antigenic variation.[291]
A histone 3 lysine 4 methyltransferase - PfSET10 - which localizes exclusively to the perinuclear active var gene expression site and is required to maintain the active var gene in a poised state during division for reactivation in daughter parasites.[631]
The variant silencing SET gene is an analog of the Drosophila melanogaster ASH1.[107] It controls histone H3 lysine 36 trimethylation (H3K36me3) on the var genes. Knocking this gene out results in the transcription of virtually all the var genes in the genome and their subsequent translation and localization to the cell membrane. This protein is present along the entire gene body including the intronic promoter and the transcription start site.
The origin recognition complex 1 protein binds the telomeres via its N terminal and appears to play some role in gene regulation.[632]
Within the regulatory region of the var genes there are a number of insulator like elements.[633] These along with the strict pairing of the 5' promoter with the second promoter within the intron are essential for the normal regulation of the var genes.
There is a small upstream (5') open reading frame associated with the var2csa gene.[634] This gene is only expressed in placental malaria. The product of the open reading frame interacts with the sequence surrounding the var2csa gene to control its transcription.
Expression of gene appears to be non random with a global activation hierarchy favouring short and highly diverse genes in central chromosomal location.[635] Longer and more conserved genes are rarely activated.
- Molecular biology
The intracellular portion of the EMP1 protein binds to PFI1780w - a member of the Plasmodium helical interspersed sub-telomeric (PHIST) family.[636]
A low level solution of the ~300 kiloDalton ectodomain of PfEMP1 has been reported.[637] It is rigid, elongated and monomeric and interacts with intercellular adhesion molecule 1 through the Duffy binding Lβ domain alone, forming a 1:1 complex.
The strength of adhesion between the EMP1 and chondroitin sulfate A is decreased at 40C compared with its binding at 37C.[638]
The subdomain 3 which connects the h6 and h7 α-helices of PfEMP1-DBL1α appears to be important in the process of parasitized erythrocytes forming rosettes with uninfected erythrocytes.[639]
- Clinical notes
It is thought that associations may exist between different var types and the clinical syndromes seen in malaria. The VAR2CSA variants bind to glycosaminoglycan chondroitin-sulfate A and have been associated with placental malaria.[640] Group A and specifically Domanin Casette type 8 have been associated with cerebral malaria in Benin.[641] Severe childhood malaria is associated with expression of specific PfEMP1 subtypes containing domain cassettes 8 and 13. These subtypes bind the endothelial protein C receptor (EPRC) with their amino terminal cysteine rich interdomain region.[642] This binding interferes with protein C binding to EPCR.
A difference between homology blocks has been noted in the phenotypes of erythrocyte rosetting and impaired consciousness.[643] The clinical significance - if any - of this finding is not presently clear.
rif family
The rif genes encode for repetitive interspersed family (rifin) proteins. The genes are found in the subtelomeric regions of the chromosomes. There exist an estimated 149 rif genes within the genome.[3]
The rifins are divided into 2 types (A and B) depending on the presence/absence of a 25 amino acid domain in the semiconserved domain.[644] One of the rifins (PF13_0006) is transcribed both in the sexual and asexual stages. It is present on both gametocytes and merozoites.
Rifin proteins are ultimately transported to the erythrocyte membrane. The function of these proteins is currently unknown.
stevor family
The stevor genes encode for the sub-telomeric variable open reading frame (stevor) proteins. The genes are found in the subtelomeric regions of the chromosomes. There exist an estimated 28 stevor genes within the genome.[3]
These proteins appear to affect membrane deformability.[645]
References
- ^ Wirth, Dyann (3 October 2002). "The parasite genome: Biological revelations". Nature. 419 (6906): 495–496. doi:10.1038/419495a. PMID 12368862.
- ^ "DPDx - Malaria Image Library".
- ^ a b c d e f g h i j k l m n o p q r s t u v w x Gardner, Malcolm; Hall, N; Fung, E; White, O; Berriman, M; Hyman, RW; Carlton, JM; Pain, A; Nelson, KE (3 October 2002). "Genome sequence of the human malaria parasite Plasmodium falciparum". Nature. 419 (6906): 498–511. doi:10.1038/nature01097. PMID 12368864.
{{cite journal}}:|first10=missing|last10=(help);|first11=missing|last11=(help);|first12=missing|last12=(help);|first13=missing|last13=(help);|first14=missing|last14=(help);|first15=missing|last15=(help);|first16=missing|last16=(help);|first17=missing|last17=(help);|first18=missing|last18=(help);|first19=missing|last19=(help);|first20=missing|last20=(help);|first21=missing|last21=(help);|first22=missing|last22=(help);|first23=missing|last23=(help);|first24=missing|last24=(help);|first25=missing|last25=(help);|first26=missing|last26=(help);|first27=missing|last27=(help);|first28=missing|last28=(help);|first29=missing|last29=(help);|first30=missing|last30=(help); Unknown parameter|displayauthors=ignored (|display-authors=suggested) (help) - ^ Jiang H, Li N, Gopalan V, Zilversmit MM, Varma S, Nagarajan V, Li J, Mu J, Hayton K, Henschen B, Yi M, Stephens R, McVean G, Awadalla P, Wellems TE, Su XZ (2011) High recombination rates and hotspots in a Plasmodium falciparum genetic cross" Genome Biol 12(4) R33
- ^ Pryde FE, Gorham HC, Louis EJ (1997) Chromosome ends: all the same under their caps. Curr Opin Genet Dev 7(6) 822-828
- ^ Stavenhagen JB, Zakian VA (1994). "Internal tracts of telomeric DNA act as silencers in Saccharomyces cerevisiae". Genes Dev. 8 (12): 1411–1422. doi:10.1101/gad.8.12.1411. PMID 7926741.
- ^ Barry JD, Ginger ML, Burton P, McCulloch R (2003) Why are parasite contingency genes often associated with telomeres? Int J Parasitol 33(1) 29-45
- ^ Freitas-Junior LH; Bottius E; Pirrit LA; Deitsch KW; Scheidig C; Guinet F; Nehrbass U; Wellems TE; Scherf A (2000). "Frequent ectopic recombination of virulence factor genes in telomeric chromosome clusters of P. falciparum". Nature. 407 (6807): 1018–1022. doi:10.1038/35039531. PMID 11069183.
{{cite journal}}: Invalid|display-authors=9(help); Unknown parameter|author-separator=ignored (help) - ^ a b c d Bozdech, Zbynek; Llinás, Manuel; Pulliam, Brian Lee; Wong, Edith D.; Zhu, Jingchun; Derisi, Joseph L. (August 18, 2003). "The Transcriptome of the Intraerythrocytic Developmental Cycle of Plasmodium falciparum". PLoS Biology. 1 (1): E5. doi:10.1371/journal.pbio.0000005. PMC 176545. PMID 12929205.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Ponts N, Harris EY, Prudhomme J, Wick I, Eckhardt-Ludka C, Hicks GR, Hardiman G, Lonardi S, Le Roch KG. Nucleosome landscape and control of transcription in the human malaria parasite. Genome Res.
- ^ a b Zhang X, Tolzmann CA, Melcher M, Haas BJ, Gardner MJ, Smith JD, Feagin JE (2011). "Branch point identification and sequence requirements for intron splicing in Plasmodium falciparum". Eukaryotic Cell. 10 (11): 1422. doi:10.1128/EC.05193-11.
{{cite journal}}: CS1 maint: multiple names: authors list (link) Cite error: The named reference "Zhang2011" was defined multiple times with different content (see the help page). - ^ a b c Florens, Laurence; Washburn, Michael P.; Raine, J. Dale; Anthony, Robert M.; Grainger, Munira; Haynes, J. David; Moch, J. Kathleen; Muster, Nemone; Sacci, John B.; Tabb, David L.; Witney, Adam A.; Wolters, Dirk; Wu, Yimin; Gardner, Malcolm J.; Holder, Anthony A.; Sinden, Robert E.; Yates, John R.; Carucci, Daniel J. (3 October 2002). "A proteomic view of the Plasmodium falciparum life cycle". Nature. 419 (6906): 520–526. doi:10.1038/nature01107. PMID 12368866.
- ^ Patakottu BR, Singh PK, Malhotra P, Chauhan VS, Patankar S (2011). "In vivo analysis of translation initiation sites in Plasmodium falciparum". Mol Biol Rep.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b "Malaria eModule - Transmission".[dead link]
- ^ a b Lindner SE, Mikolajczak SA, Vaughan AM, Moon W, Joyce BR, Sullivan WJ Jr, Kappe SH (2013) Perturbations of Plasmodium Puf2 expression and RNA-seq of Puf2-Deficient sporozoites reveal a critical role in maintaining RNA homeostasis and parasite transmissibility. Cell Microbiol. 2013 Jan 29. doi:10.1111/cmi.12116 Cite error: The named reference "Lindner2013" was defined multiple times with different content (see the help page).
- ^ "Malaria Site: Anopheles Mosquito".
- ^ Figueiredo AC, de Sanctis D, Gutiérrez-Gallego R, Cereija TB, Macedo-Ribeiro S, Fuentes-Prior P, Pereira PJ (2012) Unique thrombin inhibition mechanism by anophelin, an anticoagulant from the malaria vector. Proc Natl Acad Sci USA
- ^ Tavares J, Formaglio P, Thiberge S, Mordelet E, Van Rooijen N, Medvinsky A, Ménard R, Amino R (2013) Role of host cell traversal by the malaria sporozoite during liver infection. J Exp Med
- ^ "Malaria eModule - Exo-Erythrocytic Stages".[dead link]
- ^ a b Arévalo-Pinzón G, Curtidor H, Muñoz M, Patarroyo MA, Patarroyo ME (2011). "Synthetic peptides from two Pf sporozoite invasion-associated proteins specifically interact with HeLa and HepG2 cells". Peptides. 32 (9): 1902–8. doi:10.1016/j.peptides.2011.08.008. PMID 21864602.
{{cite journal}}: CS1 maint: multiple names: authors list (link) Cite error: The named reference "Arévalo-Pinzón2011" was defined multiple times with different content (see the help page). - ^ Eaton P, Zuzarte-Luis V, Mota MM, Santos NC, Prudêncio M (2011). "Infection by Plasmodium changes shape and stiffness of hepatic cells". Nanomedicine. 8 (1): 17–9. doi:10.1016/j.nano.2011.10.004. PMID 22033078.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Coppens I (2011). "Metamorphoses of malaria: the role of autophagy in parasite differentiation". Essays Biochem. 51: 127–36. doi:10.1042/bse0510127. PMID 22023446.
- ^ Stanway RR, Mueller N, Zobiak B; et al. (2011). "Organelle segregation into Plasmodium liver stage merozoites". Cell. Microbiol. 13 (11): 1768–82. doi:10.1111/j.1462-5822.2011.01657.x. PMID 21801293.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Pei Y, Tarun AS, Vaughan AM, Herman RW, Soliman JM, Erickson-Wayman A, Kappe SH. (2010). "Plasmodium pyruvate dehydrogenase activity is only essential for the parasite's progression from liver infection to blood infection". Microbiol. 75 (4): 957–71. doi:10.1111/j.1365-2958.2009.07034.x. PMID 20487290.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Labaied M, Jayabalasingham B, Bano N, Cha SJ, Sandoval J, Guan G, Coppens I (2010). "Plasmodium salvages cholesterol internalized by LDL and synthesized de novo in the liver". Cell Microbiol. 13 (4): 569–86. doi:10.1111/j.1462-5822.2010.01555.x. PMID 21105984.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Duarte J, Herbert F, Guiyedi V, Franetich JF, Roland J, Cazenave PA, Mazier D, Kombila M, Fesel C, Pied S (2012) High levels of IgE autoantibody to 14-3-3 epsilon protein correlate with protection against severe Plasmodium falciparum malaria. J Infect Dis
- ^ Yalaoui S, Zougbédé S, Charrin S, Silvie O, Arduise C, Farhati K, Boucheix C, Mazier D, Rubinstein E, Froissard P (2008). Mota, Maria M (ed.). "Hepatocyte permissiveness to Plasmodium infection is conveyed by a short and structurally conserved region of the CD81 large extracellular domain". PLoS Pathogens. 4 (2): e1000010. doi:10.1371/journal.ppat.1000010. PMC 2279262. PMID 18389082.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Silvie O, Rubinstein E, Franetich JF, Prenant M, Belnoue E, Rénia L, Hannoun L, Eling W, Levy S, Boucheix C, Mazier D (2003). "Hepatocyte CD81 is required for Plasmodium falciparum and Plasmodium yoelii sporozoite infectivity". Nat. Med. 9 (1): 93–6. doi:10.1038/nm808. PMID 12483205.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Favretto F, Assfalg M, Molinari H, D'Onofrio M (2012) Evidence from NMR interaction studies challenges the hypothesis of direct lipid transfer from L-FABP to malaria sporozoite protein UIS3. Protein Sci. 2012 Nov 20. doi:10.1002/pro.2194
- ^ a b Müller K, Matuschewski K, Silvie O (2011). Gruner, Anne Charlotte (ed.). "The Puf-family RNA-binding protein Puf2 controls sporozoite conversion to liver stages in the malaria parasite". PLoS ONE. 6 (5): e19860. doi:10.1371/journal.pone.0019860. PMC 3097211. PMID 21673790.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Gomes-Santos CS; Braks J; Prudêncio M; Carret C; Gomes AR; Pain A; Feltwell T; Khan S; Waters A (2011). Soldati-Favre, Dominique (ed.). "Transition of Plasmodium sporozoites into liver stage-like forms is regulated by the RNA binding protein pumilio". PLoS Pathog. 7 (5): e1002046. doi:10.1371/journal.ppat.1002046. PMC 3098293. PMID 21625527.
{{cite journal}}:|first10=missing|last10=(help);|first11=missing|last11=(help); Invalid|display-authors=9(help); Unknown parameter|author-separator=ignored (help)CS1 maint: unflagged free DOI (link) - ^ Yu M, Kumar T. R, Nkrumah L. J, Coppi A, Retzlaff S; et al. (2008). "The fatty acid biosynthesis enzyme FabI plays a key role in the development of liver-stage malarial parasites". Cell Host & Microbe. 4 (6): 567–578. doi:10.1016/j.chom.2008.11.001. PMC 2646117. PMID 19064257.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ a b Portugal S; Carret C; Recker M; Armitage AE; Gonçalves LA; Epiphanio S; Sullivan D; Roy C; Newbold CI (2011). "Host-mediated regulation of superinfection in malaria". Nat Med. 17 (6): 732–737. doi:10.1038/nm.2368. PMID 21572427.
{{cite journal}}:|first10=missing|last10=(help);|first11=missing|last11=(help); Invalid|display-authors=9(help); Unknown parameter|author-separator=ignored (help) Cite error: The named reference "Portugal2011" was defined multiple times with different content (see the help page). - ^ de Mast Q, Nadjm B, Reyburn H; et al. (2009). "Assessment of urinary concentrations of hepcidin provides novel insight into disturbances in iron homeostasis during malarial infection". J. Infect. Dis. 199 (2): 253–62. doi:10.1086/595790. PMID 19032104.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Wang HZ, He YX, Yang CJ, Zhou W, Zou CG (2011). "Hepcidin is regulated during blood-stage malaria and plays a protective role in malaria infection". J Immunol. 187 (12): 6410–6. doi:10.4049/jimmunol.1101436. PMID 22084434.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Zhao H, Konishi A, Fujita Y, Yagi M, Ohata K, Aoshi T, Itagaki S, Sato S, Narita H, Abdelgelil NH, Inoue M, Culleton R, Kaneko O, Nakagawa A, Horii T, Akira S, Ishii KJ, Coban C (2012) Lipocalin 2 bolsters innate and adaptive immune responses to blood-stage malaria infection by reinforcing host iron metabolism. Cell Host Microbe 12(5) 705-16. doi:10.1016/j.chom.2012.10.010
- ^ Gozzelino R, Andrade BB, Larsen R, Luz NF, Vanoaica L, Seixas E, Coutinho A, Cardoso S, Rebelo S, Poli M, Barral-Netto M, Darshan D, Kühn LC, Soares MP (2012) Metabolic adaptation to tissue iron overload confers tolerance to malaria. Cell Host Microbe 12(5) 693-704. doi:10.1016/j.chom.2012.10.011
- ^ a b c Giovannini D, Späth S, Lacroix C; et al. (2011). "Independent roles of apical membrane antigen 1 and rhoptry neck proteins during host cell invasion by apicomplexa". Cell Host Microbe. 10 (6): 591–602. doi:10.1016/j.chom.2011.10.012. PMID 22177563.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) Cite error: The named reference "Giovannini2011" was defined multiple times with different content (see the help page). - ^ Gomes-Santos CS, Itoe MA, Afonso C; et al. (2012). Kappe, Stefan (ed.). "Highly Dynamic Host Actin Reorganization around Developing Plasmodium Inside Hepatocytes". PLoS ONE. 7 (1): e29408. doi:10.1371/journal.pone.0029408. PMC 3253080. PMID 22238609.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Miller JL, Harupa A, Kappe SH, Mikolajczak SA (2012). "Plasmodium macrophage migration inhibitory factor is necessary for efficient liver stage development". Infect Immun. 80 (4): 1399–407. doi:10.1128/IAI.05861-11. PMC 3318411. PMID 22252874.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Orito Y, Ishino T, Iwanaga S, Kaneko I, Kato T, Menard R, Chinzei Y, Yuda M (2012) Liver-specific protein 2: a Plasmodium protein exported to the hepatocyte cytoplasm and required for merozoite formation. Mol Microbiol doi:10.1111/mmi.12083
- ^ Ploemen IH, Croes HJ, van Gemert GJ, Wijers-Rouw M, Hermsen CC, Sauerwein RW (2012) Plasmodium berghei Δp52&p36 Parasites develop independent of a parasitophorous vacuole membrane in Huh-7 liver cells" PLoS One 7(12) e50772. doi:10.1371/journal.pone.0050772
- ^ Kaushansky A, Metzger PG, Douglass AN, Mikolajczak SA, Lakshmanan V, Kain HS, Kappe SH (2013) Malaria parasite liver stages render host hepatocytes susceptible to mitochondria-initiated apoptosis. Cell Death Dis 4:e762. doi:10.1038/cddis.2013.286
- ^ Tokumasu F, Ostera GR, Amaratunga C, Fairhurst RM (2012) Modifications in erythrocyte membrane zeta potential by Plasmodium falciparum infection. Exp Parasitol
- ^ Quadt KA, Barfod L, Andersen D, Bruun J, Gyan B, Hassenkam T, Ofori MF, Hviid L (2012) The density of knobs on Plasmodium falciparum-infected erythrocytes depends on developmental age and varies among isolates" PLoS One 7(9) e45658. doi:10.1371/journal.pone.0045658
- ^ Butzloff S, Groves MR, Wrenger C, Müller IB (2012) Cytometric quantification of singlet oxygen in the human malaria parasite Plasmodium falciparum. Cytometry A doi:10.1002/cyto.a.22081.
- ^ Murray MC, Perkins ME (1989) Phosphorylation of erythrocyte membrane and cytoskeleton proteins in cells infected with Plasmodium falciparum. Mol Biochem Parasitol 34(3) 229-236
- ^ Pantaleo A, Ferru E, Carta F, Mannu F, Giribaldi G, Vono R, Lepedda AJ, Pippia P, Turrini F (2010) Analysis of changes in tyrosine and serine phosphorylation of red cell membrane proteins induced by P. falciparum growth" Proteomics 10(19) 3469-3479
- ^ a b Balaji SN, Trivedi V (2013) Extracellular methemoglobin primes red blood cell aggregation in malaria: An in vitro mechanistic study. FEBS Lett pii: S0014-5793(13)00003-3. doi:10.1016/j.febslet.2012.12.015 Cite error: The named reference "Balaji2013" was defined multiple times with different content (see the help page).
- ^ D'Alessandro S, Basilico N, Prato M (2013) Effects of Plasmodium falciparum-infected erythrocytes on matrix metalloproteinase-9 regulation in human microvascular endothelial cells. Asian Pac J Trop Med 6(3) 195-199 doi:10.1016/S1995-7645(13)60022-X
- ^ Shi H, Liu Z, Li A, Yin J, Chong AG, Tan KS, Zhang Y, Lim CT (2013) Life cycle-dependent cytoskeletal modifications in Plasmodium falciparum infected erythrocytes" PLoS One 8(4) e61170. doi:10.1371/journal.pone.0061170
- ^ Karimi A, Navidbakhsh M, Motevalli Haghi A, Faghihi S (2013) An innovative shape equation to quantify the morphological characteristics of parasitized red blood cells by Plasmodium falciparum and Plasmodium vivax. Proc Inst Mech Eng H 227(4) 428-37. doi:10.1177/0954411912474611
- ^ a b Hopkins J, Fowler R, Krishna S, Wilson I, Mitchell G, Bannister L (1999) The plastid in Plasmodium falciparum asexual blood stages: a three-dimensional ultrastructural analysis. Protista 150:283–295 Cite error: The named reference "Hopkins1999" was defined multiple times with different content (see the help page).
- ^ Bannister LH, Hopkins JM, Dluzewski AR, Margos G, Williams IT, Blackman MJ, Kocken CH, Thomas AW, Mitchell GH (2003) Plasmodium falciparum apical membrane antigen 1 (PfAMA-1) is translocated within micronemes along subpellicular microtubules during merozoite development" J Cell Sci 116(18) 3825-3834
- ^ Wickramarachchi T, Devi YS, Mohmmed A, Chauhan VS (2013) Identification and characterization of a novel Plasmodium falciparum merozoite apical protein involved in erythrocyte binding and invasion" PLoS One 3(3) e1732. doi:10.1371/journal.pone.0001732
- ^ a b Hans N, Singh S, Jain SK, Chauhan VS (2013) Identification of novel rhoptry neck protein of Plasmodium falciparum. Mol Biochem Parasitol pii: S0166-6851(13)00032-7. doi:10.1016/j.molbiopara.2013.02.007 Cite error: The named reference "Hans2013" was defined multiple times with different content (see the help page).
- ^ Yahata K, Treeck M, Culleton R, Gilberger TW, Kaneko O (2012) Time-lapse imaging of red blood cell invasion by the rodent malaria parasite Plasmodium yoelii" PLoS One 7(12) e50780. doi:10.1371/journal.pone.0050780
- ^ a b Cowman, AF; Crabb, BS (24 February 2006). "Invasion of Red Blood Cells by Malaria Parasites". Cell. 124 (4): 755–766. doi:10.1016/j.cell.2006.02.006. PMID 16497586.
- ^ Chenniappan K (2011). "Alternative pathways of erythrocyte invasion, parasite multiplication potential and severity of the clinical episode of P. falciparum malaria in the Peruvian Amazon". Parasitol Res. 110 (2): 1019. doi:10.1007/s00436-011-2663-2. PMID 21993880.
- ^ Huang X, Liew K, Natalang O, Siau A, Zhang N, Preiser PR (2013) The role of serine-type serine repeat antigen in Plasmodium yoelii blood stage development" PLoS One 8(4) e60723. doi:10.1371/journal.pone.0060723
- ^ a b Miao J, Wang Z, Liu M, Parker D, Li X, Chen X, Cui L (2013) Plasmodium falciparum: Generation of pure gametocyte culture by heparin treatment. Exp Parasitol pii: S0014-4894(13)00248-8. doi:10.1016/j.exppara.2013.09.010 Cite error: The named reference "Miao2013" was defined multiple times with different content (see the help page).
- ^ Kobayashi K, Takano R, Takemae H, Sugi T, Ishiwa A, Gong H, Recuenco FC, Iwanaga T, Horimoto T, Akashi H, Kato K (2013) Analyses of interactions between heparin and the apical ssurface proteins of Plasmodium falciparum. Sci Rep 3:3178. doi: 10.1038/srep03178
- ^ a b c Boyle MJ, Richards JS, Gilson PR, Chai W, Beeson JG (2010) Interactions with heparin-like molecules during erythrocyte invasion by Plasmodium falciparum merozoites. Blood 115(22):4559-4568. doi: 10.1182/blood-2009-09-243725 Cite error: The named reference "Boyle2010" was defined multiple times with different content (see the help page).
- ^ Bozdech Z, Llinas M, Pulliam BL, Wong ED, Zhu J, et al (2003) The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol 1:E5
- ^ a b Singh, S; Alam, MM; Pal-Bhowmick, I; Brzostowski, JA; Chitnis, CE (2010). Blackman, Michael John (ed.). "Distinct external signals trigger sequential release of apical organelles during erythrocyte invasion by malaria parasites". PLoS Pathog. 6 (2): e1000746. doi:10.1371/journal.ppat.1000746.
{{cite journal}}: Unknown parameter|author-separator=ignored (help)CS1 maint: unflagged free DOI (link) Cite error: The named reference "Singh2010" was defined multiple times with different content (see the help page). - ^ Bansal A, Singh S, More KR, Hans D, Nangalia K, Yogavel M, Sharma A, Chitnis CE (2012) Characterization of Plasmodium falciparum calcium dependent protein kinase 1 (PfCDPK1) and its role in microneme secretion during erythrocyte invasion. J Biol Chem
- ^ a b Singh S, More KR, Chitnis CE (2013) Role of calcineurin and actin dynamics in regulated secretion of microneme proteins in Plasmodium falciparum merozoites during erythrocyte invasion. Cell Microbiol doi:10.1111/cmi.12177 Cite error: The named reference "Singh2013" was defined multiple times with different content (see the help page).
- ^ Siddiqui FA, Dhawan S, Singh S, Singh B, Gupta P, Pandey A, Mohmmed A, Gaur D, Chitnis CE (2013) A thrombospondin structural repeat containing rhoptry protein from Plasmodium falciparum mediates erythrocyte invasion. Cell Microbiol doi:10.1111/cmi.12118
- ^ Thomas, DC; Ahmed, A; Gilberger, TW; Sharma, P (2012). Blader, Ira (ed.). "Regulation of Plasmodium falciparum Glideosome Associated Protein 45 (PfGAP45) Phosphorylation". PLoS ONE. 7 (4): e35855. doi:10.1371/journal.pone.0035855.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Ord RL, Rodriguez M, Yamasaki T, Takeo S, Tsuboi T, Lobo CA (2012). Templeton, Thomas J (ed.). "Targeting Sialic Acid Dependent and Independent Pathways of Invasion in Plasmodium falciparum". PLoS ONE. 7 (1): e30251. doi:10.1371/journal.pone.0030251. PMC 3257272. PMID 22253925.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Cowman AF, Crabb BS (2006) Invasion of red blood cells by malaria parasites. Cell 124:755–766
- ^ Rydzak J, Kryńska K, Suchanowska A, Kaczmarek R, Lukasiewicz J, Czerwiński M, Jaśkiewicz E (2012) Bacterially expressed truncated F2 domain of Plasmodium falciparum EBA-140 antigen can bind to human erythrocytes. Acta Biochim Pol
- ^ Sim BKL, Chitnis CE, Wasniowska K, Hadley TJ, Miller LH (1994) Receptor and ligand domains for invasion of erythrocytes by Plasmodium falciparum. Science 264:1941–1944
- ^ Mayer DC et al (2009) Glycophorin B is the erythrocyte receptor of Plasmodium falciparum erythrocyte-binding ligand, EBL-1. Proc Natl Acad Sci USA 106:5348–5352
- ^ Maier AG et al (2003) Plasmodium falciparum erythrocyte invasion through glycophorin C and selection for Gerbich negativity in human populations. Nat Med 9:87–92
- ^ Li X, Marinkovic M, Russo C, McKnight CJ, Coetzer TL, Chishti AH (2012). "Identification of a specific region of Plasmodium falciparum EBL-1 that binds to host receptor glycophorin B and inhibits merozoite invasion in human red blood cells". Mol Biochem Parasitol. 183 (1): 23–31. doi:10.1016/j.molbiopara.2012.01.002. PMC 3307866. PMID 22273481.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Adams JH, Sim BK, Dolan SA, Fang X, Kaslow DC, Miller LH (1992) A family of erythrocyte binding proteins of malaria parasites Proc. Natl Acad Sci USA 89:7085-7089
- ^ Ambroggio X, Jiang L, Aebig J, Obiakor H, Lukszo J, Narum DL (2013) The epitope of monoclonal antibodies blocking erythrocyte invasion by Plasmodium falciparum map to The dimerization and receptor glycan binding Sites of EBA-175" PLoS One 8(2) e56326. doi:10.1371/journal.pone.0056326
- ^ Lin DH, Malpede BM, Batchelor JD, Tolia NH (2012) Crystal and solution structures of Plasmodium falciparum erythrocyte binding antigen 140 reveal determinants of receptor specificity during erythrocyte invasion. J Biol Chem
- ^ Wanaguru MK, Crosnier C, Johnson S, Rayner JC, Wright GJ (2013) A biochemical analysis of the Plasmodium falciparum erythrocyte binding antigen-175 (EBA175) - glycophorin-A interaction: implications for vaccine design. J Biol Chem
- ^ Martinez C, Marzec T, Smith CD, Tell LA, Sehgal RN (2012) Identification and expression of maebl, an erythrocyte-binding gene, in Plasmodium gallinaceum. Parasitol Res
- ^ Arumugam TU, Takeo S, Yamasaki T, Thonkukiatkul A, Miura K, Otsuki H, Zhou H, Long CA, Sattabongkot J, Thompson J, Wilson DW, Beeson JG, Healer J, Crabb BS, Cowman AF, Torii M, Tsuboi T (2011). "Discovery of GAMA, a Plasmodium falciparum merozoite micronemal protein, as a novel blood-stage vaccine candidate antigen". Infect Immun. 79 (11): 4523–32. doi:10.1128/IAI.05412-11. PMC 3257921. PMID 21896773.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Taylor HM; Triglia T; Thompson J; Sajid M; Fowler R; et al. (2001). "Plasmodium falciparum homologue of the genes for Plasmodium vivax and Plasmodium yoelii adhesive proteins, which is transcribed but not translated". Infect Immun. 69 (6): 3635–3645. doi:10.1128/IAI.69.6.3635-3645.2001. PMC 98354. PMID 11349024.
{{cite journal}}: Unknown parameter|author-separator=ignored (help) - ^ Rayner JC, Vargas-Serrato E, Huber CS, Galinski MR, Barnwell JW (2001) A Plasmodium falciparum homologue of Plasmodium vivax reticulocyte binding protein (PvRBP1) defines a trypsin-resistant erythrocyte invasion pathway. J Exp Med 194:1571–1581
- ^ Gao X, Yeo KP, Aw SS, Kuss C, Iyer JK, Genesan S, Rajamanonmani R, Lescar J, Bozdech Z, Preiser PR (2008) Antibodies targeting the PfRH1 binding domain inhibit invasion of Plasmodium falciparum merozoites. PLoS Pathog 4:e1000104 doi:10.1371/journal.ppat.1000104
- ^ Duraisingh MT, Triglia T, Ralph SA, Rayner JC, Barnwell JW, McFadden GI, Cowman AF (2003) Phenotypic variation of Plasmodium falciparum merozoite proteins directs receptor targeting for invasion of human erythrocytes. EMBO J 22:1047–1057
- ^ Dvorin JD, Bei AK, Coleman BI, Duraisingh MT (2010). "Functional diversification between two related Plasmodium falciparum merozoite invasion ligands is determined by changes in the cytoplasmic domain". Mol Microbiol. 75 (4): 990–1006. doi:10.1111/j.1365-2958.2009.07040.x. PMC 3627358. PMID 20487292.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Gunalan K, Gao X, Liew KJ, Preiser PR (2011). "Differences in erythrocyte receptor specificity of different parts of the Plasmodium falciparum reticulocyte binding protein homologue 2a". Infect Immun. 79 (8): 3421–30. doi:10.1128/IAI.00201-11. PMC 3147545. PMID 21628513.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Triglia T; Chen L; Lopaticki S; Dekiwadia C; Riglar DT; Hodder AN; Ralph SA; Baum J; Cowman AF (2011). Kazura, James W (ed.). "Plasmodium falciparum merozoite invasion is inhibited by antibodies that target the PfRh2a and b binding domains". PLoS Pathog. 7 (6): e1002075. doi:10.1371/journal.ppat.1002075.
{{cite journal}}: Invalid|display-authors=9(help); Unknown parameter|author-separator=ignored (help)CS1 maint: unflagged free DOI (link) - ^ Baum J, Maier AG, Good RT, Simpson KM, Cowman AF (2005) Invasion by P. falciparum merozoites suggests a hierarchy of molecular interactions" PLoS Pathog 1(4) e37
- ^ Park HJ, Guariento M, Maciejewski M, Hauhart R, Tham WH, Cowman AF, Schmidt CQ, Mertens HD, Liszewski MK, Hourcade DE, Barlow PN, Atkinson JP (2013) Using mutagenesis and structural biology to map the binding site for the Plasmodium falciparum merozoite protein PfRh4 on the human immune adherence receptor. J Biol Chem
- ^ Tham WH et al (2009) Antibodies to reticulocyte-binding protein-like homologue 4 inhibit invasion of Plasmodium falciparum into human erythrocytes. Infect Immun 77:2427–2435
- ^ Chen L; Lopaticki S; Riglar DT; Dekiwadia C; Uboldi AD; Tham WH; O'Neill MT; Richard D; Baum J (2011). Blackman, Mike John (ed.). "An EGF-like protein forms a complex with PfRh5 and is required for invasion of human erythrocytes by Plasmodium falciparum". PLoS Pathog. 7 (9): e1002199. doi:10.1371/journal.ppat.1002199. PMC 3164636. PMID 21909261.
{{cite journal}}:|first10=missing|last10=(help);|first11=missing|last11=(help); Invalid|display-authors=9(help); Unknown parameter|author-separator=ignored (help)CS1 maint: unflagged free DOI (link) - ^ a b Baum J, Chen L, Healer J; et al. (2009). "Reticulocyte-binding protein homologue 5 - an essential adhesin involved in invasion of human erythrocytes by Plasmodium falciparum". Int. J. Parasitol. 39 (3): 371–80. doi:10.1016/j.ijpara.2008.10.006. PMID 19000690.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) Cite error: The named reference "Baum2009" was defined multiple times with different content (see the help page). - ^ Crosnier C, Bustamante LY, Bartholdson SJ; et al. (2011). "Basigin is a receptor essential for erythrocyte invasion by Plasmodium falciparum". Nature. 480 (7378): 534–7. doi:10.1038/nature10606. PMC 3245779. PMID 22080952.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Tran TM, Ongoiba A, Coursen J, Crosnier C, Diouf A, Huang CY, Li S, Doumbo S, Doumtabe D, Kone Y, Bathily A, Dia S, Niangaly M, Dara C, Sangala J, Miller LH, Doumbo OK, Kayentao K, Long CA, Miura K, Wright GJ, Traore B, Crompton PD (2013) Naturally acquired antibodies specific for Plasmodium falciparum RH5 inhibit parasite growth and predict protection from malaria. J Infect Dis
- ^ Lopaticki S; Maier AG; Thompson J; Wilson DW; Tham WH; et al. (2011). "Reticulocyte and erythrocyte binding-like proteins function cooperatively in invasion of human erythrocytes by malaria parasites". Infect Immun. 79 (3): 1107–1117. doi:10.1128/IAI.01021-10. PMC 3067488. PMID 21149582.
{{cite journal}}:|first10=missing|last10=(help);|first11=missing|last11=(help); Unknown parameter|author-separator=ignored (help) - ^ Wickramarachchi T, Devi YS, Mohmmed A, Chauhan VS (2008) Identification and characterization of a novel Plasmodium falciparum merozoite apical protein involved in erythrocyte binding and invasion. PLoS One 3(3):e1732. doi: 10.1371/journal.pone.0001732
- ^ Alexander DL, Arastu-Kapur S, Dubremetz JF, Boothroyd JC (2006) Plasmodium falciparum AMA1 binds a rhoptry neck protein homologous to TgRON4, a component of the moving junction in Toxoplasma gondii. Eukaryot Cell 5:1169–1173.
- ^ Clark JT, Anand R, Akoglu T, McBride JS (1987) Identification and characterisation of proteins associated with the rhoptry organelles of Plasmodium falciparum merozoites. Parasitol Res 73:425–434
- ^ Topolska AE, Lidgett A, Truman D, Fujioka H, Coppel RL (2004) Characterization of a membrane-associated rhoptry protein of Plasmodium falciparum. J Biol Chem 279:4648–4656
- ^ Ling IT, Florens L, Dluzewski AR, Kaneko O, Grainger M, et al (2004) The Plasmodium falciparum clag9 gene encodes a rhoptry protein that is transferred to the host erythrocyte upon invasion. Mol Microbiol 52:107–118
- ^ Hiller NL, Akompong T, Morrow JS, Holder AA, Haldar K (2003) Identification of a stomatin orthologue in vacuoles induced in human erythrocytes by malaria parasites: A role for microbial raft proteins in Apicomplexan vacuole biogenesis. J Biol Chem 278:48413–48421
- ^ Peterson MG, Marshall VM, Smythe JA, Crewther PE, Lew A, Silva A, Anders RF, Kemp DJ (1989) Integral membrane protein located in the apical complex of Plasmodium falciparum. Mol Cell Biol 9:3151-3154
- ^ Holder AA, Blackman MJ, Burghaus PA, Chappel JA, Ling IT, McCallum-Deighton N, Shai S (1992) A malaria merozoite surface protein (MSP1)-structure, processing and function. Mem Inst Oswaldo Cruz 87 Suppl 3:37-42
- ^ a b Zhang X, Dong Y, Yu J, Tu X (2013) Effects of environmental factors on MSP21-25 aggregation indicate the roles of hydrophobic and electrostatic interactions in the aggregation process. Eur Biophys J Cite error: The named reference "Zhang2013" was defined multiple times with different content (see the help page).
- ^ a b Jiang J, Barnwell JW, Meyer EV, Galinski MR. Plasmodium vivax merozoite surface protein-3 (PvMSP3) expression of an 11 member multigene family in blood-stage parasites" PLoS One 8(5) e63888. doi:10.1371/journal.pone.0063888 Cite error: The named reference "Jiang2013" was defined multiple times with different content (see the help page).
- ^ Black CG, Wu T, Wang L, Hibbs AR, Coppel RL (2001) Merozoite surface protein 8 of Plasmodium falciparum contains two epidermal growth factor-like domains. Mol Biochem Parasitol 114(2):217-226
- ^ Chenet SM, Pacheco MA, Bacon DJ, Collins WE, Barnwell JW, Escalante AA (2013) The evolution and diversity of a low complexity vaccine candidate, merozoite surface protein 9 (MSP-9), in Plasmodium vivax and closely related species. Infect Genet Evol 20C:239-248. doi:10.1016/j.meegid.2013.09.011
- ^ Andreína Pacheco M, Elango AP, Rahman AA, Fisher D, Collins WE, Barnwell JW, Escalante AA (2012) Evidence of purifying selection on merozoite surface protein 8 (MSP8) and 10 (MSP10) in Plasmodium spp. Infect Genet Evol
- ^ Bartholdson SJ, Bustamante LY, Crosnier C, Johnson S, Lea S, Rayner JC, Wright GJ (2012) Semaphorin-7A is an erythrocyte receptor for P. falciparum merozoite-specific TRAP homolog, MTRAP" PLoS Pathog 8(11) e1003031. doi:10.1371/journal.ppat.1003031
- ^ Bartholdson SJ, Crosnier C, Bustamante LY, Rayner JC, Wright GJ (2013) Identifying novel Plasmodium falciparum erythrocyte invasion receptors using systematic extracellular protein interaction screens. Cell Microbiol doi:10.1111/cmi.12151
- ^ Pandey AK, Reddy KS, Sahar T, Gupta S, Singh H, Reddy EJ, Asad M, Siddiqui FA, Gupta P, Singh B, More KR, Mohmmed A, Chitnis CE, Chauhan VS, Gaur D (2013) Identification of a potent combination of key Plasmodium falciparum merozoite antigens that elicit strain-transcending parasite-neutralizing antibodies. Infect Immun 81(2):441-451 doi: 10.1128/IAI.01107-12
- ^ Pelleau S, Diop S, Badiane MD, Vitte J, Beguin P, Nato F, Diop BM, Bongrand P, Parzy D, Jambou R (2012) Enhanced basophil reactivities during severe malaria and their relationship with the plasmodial histamine releasing factor PfTCTP. Infect Immun
- ^ Dreyer AM, Matile H, Papastogiannidis P, Kamber J, Favuzza P, Voss TS, Wittlin S, Pluschke G (2012) Passive immunoprotection of Plasmodium falciparum-infected mice designates the CyRPA as candidate malaria vaccine antigen. J Immunol
- ^ Chatterjee S, Singh S, Sohoni R, Kattige V, Deshpande C, Chiplunkar S, Kumar N, Sharma S (2000) Characterization of domains of the phosphoriboprotein P0 of Plasmodium falciparum. Mol Biochem Parasitol 107(2) 143-154
- ^ Farrell A, Thirugnanam S, Lorestani A, Dvorin JD, Eidell KP, Ferguson DJ, Anderson-White BR, Duraisingh MT, Marth GT; et al. (2012). "A DOC2 protein identified by mutational profiling is essential for apicomplexan parasite exocytosis". Science. 335 (6065): 218–221. doi:10.1126/science.1210829. PMC 3354045. PMID 22246776.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Duncan RR, Shipston MJ, Chow RH (2000). "Double C2 protein. A review". Biochimie. 82 (5): 421–426. doi:10.1016/S0300-9084(00)00214-5. PMID 10865129.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Tyagi RK, Sharma YD (2012) Erythrocyte binding activity displayed by a selective group of Plasmodium vivax tryptophan rich antigens is inhibited by patients' antibodies" PLoS One 7(12) e50754. doi:10.1371/journal.pone.0050754
- ^ Moreno-Pérez DA, Saldarriaga A, Patarroyo MA (2013) Characterizing PvARP, a novel Plasmodium vivax antigen. Malar J 12:165. doi:10.1186/1475-2875-12-165
- ^ Wickramarachchi T, Cabrera AL, Sinha D, Dhawan S, Chandran T, Devi YS, Kono M, Spielmann T, Gilberger TW, Chauhan VS, Mohmmed A (2009) A novel Plasmodium falciparum erythrocyte binding protein associated with the merozoite surface, PfDBLMSP. Int J Parasitol 39(7):763-773
- ^ Curtidor H, Patiño LC, Arévalo-Pinzón G, Vanegas M, Patarroyo ME, Patarroyo MA (2013) Plasmodium falciparum rhoptry neck protein 5 peptides bind to human red blood cells and inhibit parasite invasion. Peptides pii: S0196-9781(13)00268-4. doi:10.1016/j.peptides.2013.07.028
- ^ a b Lamarque M; Besteiro S; Papoin J; Roques M; Vulliez-Le Normand B; Morlon-Guyot J; Dubremetz JF; Fauquenoy S; Tomavo S (2011). Soldati-Favre, Dominique (ed.). "The RON2-AMA1 interaction is a critical step in moving junction-dependent invasion by apicomplexan parasites". PLoS Pathog. 7 (2): e1001276. doi:10.1371/journal.ppat.1001276.
{{cite journal}}:|first10=missing|last10=(help);|first11=missing|last11=(help); Invalid|display-authors=9(help); Unknown parameter|author-separator=ignored (help)CS1 maint: unflagged free DOI (link) Cite error: The named reference "Lamarque2011" was defined multiple times with different content (see the help page). - ^ Hossain ME, Dhawan S, Mohmmed A (2011). "The cysteine-rich regions of Plasmodium falciparum RON2 bind with host erythrocyte and AMA1 during merozoite invasion". Parasitol Res. 110 (5): 1711–21. doi:10.1007/s00436-011-2690-z. PMID 22033736.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b Tonkin ML, Crawford J, Lebrun ML, Boulanger MJ (2013) Babesia divergens and Neospora caninum apical membrane antigen 1 structures reveal selectivity and plasticity in apicomplexan parasite host cell invasion" Protein Sci 22(1) 114-127 doi:10.1002/pro.2193 Cite error: The named reference "Tonkin2013" was defined multiple times with different content (see the help page).
- ^ Levano-Garcia J, Dluzewski AR, Markus RP, Garcia CR (2010). "Purinergic signalling is involved in the malaria parasite Plasmodium falciparum invasion to red blood cells". Purinergic Signal. 6 (4): 365–372. doi:10.1007/s11302-010-9202-y. PMC 3033500. PMID 21437007.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Knuepfer E, Suleyman O, Dluzewski AR, Straschil U, O'Keeffe AH, Ogun SA, Green JL, Grainger M, Tewari R, Holder AA (2013) RON12, a novel Plasmodium-specific rhoptry neck protein important for parasite proliferation. Cell Microbiol doi:10.1111/cmi.12181
- ^ Ridzuan, MA; Moon, RW; Knuepfer, E; Black, S; Holder, AA; Green, JL (2012). Langsley, Gordon (ed.). "Subcellular location, phosphorylation and assembly into the motor complex of GAP45 during Plasmodium falciparum schizont development". PLoS ONE. 7 (3): e33845. doi:10.1371/journal.pone.0033845.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Pal-Bhowmick I, Andersen J, Srinivasan P, Narum DL, Bosch J, Miller LH (2012) Binding of aldolase and glyceraldehyde-3-phosphate dehydrogenase to the cytoplasmic tails of Plasmodium falciparum merozoite Duffy binding-like and reticulocyte homology ligands. MBio. 2012 Sep 18;3(5). pii: e00292-12. doi:10.1128/mBio.00292-12
- ^ a b "Malaria eModule - ASEXUAL ERYTHROCYTIC STAGES".[dead link]
- ^ Hanssen E, Knoechel C, Dearnley M, Dixon MW, Le Gros M, Larabell C, Tilley L (2011). "Soft X-ray microscopy analysis of cell volume and hemoglobin content in erythrocytes infected with asexual and sexual stages of Plasmodium falciparum". J Struct Biol. 177 (2): 224–32. doi:10.1016/j.jsb.2011.09.003. PMC 3349340. PMID 21945653.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Fedosov DA, Lei H, Caswell B, Suresh S, Karniadakis GE (2011). Beard, Daniel A (ed.). "Multiscale modeling of red blood cell mechanics and blood flow in malaria". PLoS Comput Biol. 7 (12): e1002270. doi:10.1371/journal.pcbi.1002270. PMC 3228770. PMID 22144878.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ a b Francis, SE; Sullivan Dj, Jr; Goldberg, DE (1997). "Hemoglobin metabolism in the malaria parasite Plasmodium falciparum". Ann. Review Micro. 51 (1): 97–123. doi:10.1146/annurev.micro.51.1.97. PMID 9343345.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) Cite error: The named reference "Francis1997" was defined multiple times with different content (see the help page). - ^ Krugliak, M. (2002). "Intraerythrocytic Plasmodium falciparum utilizes only a fraction of the amino acids derived from the digestion of host cell cytosol for the biosynthesis of its proteins". Molecular and Biochemical Parasitology. 119 (2): 249–256. doi:10.1016/S0166-6851(01)00427-3. PMID 11814576.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Ginsburg, H. (1983). "New permeability pathways induced in membranes of Plasmodium falciparum". Mol. Biochem. Parasitol. 8 (2): 177–190. doi:10.1016/0166-6851(83)90008-7. PMID 6348537.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Lew, VL; Tiffert, T; Ginsburg, H (2003). "Excess hemoglobin digestion and the osmotic stability of Plasmodium falciparum-infected red blood cells". Blood. 101 (10): 4189–4194. doi:10.1182/blood-2002-08-2654. PMID 12531811.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Esposito, A; et al. (2008). Schnur, Joel M. (ed.). "FRET Imaging of Hemoglobin Concentration in Plasmodium falciparum-Infected Red Cells". PLoS ONE. 3 (11): e3780. doi:10.1371/journal.pone.0003780. PMC 2582953. PMID 19023444.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Lang F, Lang PA, Lang KS, Brand V, Tanneur V, Duranton C, Wieder T, Huber SM (2004). "Channel-induced apoptosis of infected host cells-the case of malaria". Pflugers Arch. 448 (3): 319–324. doi:10.1007/s00424-004-1254-9. PMID 15042371.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Margos G, Bannister LH, Dluzewski AR, Hopkins J, Williams IT, Mitchell GH (2004) Correlation of structural development and differential expression of invasion-related molecules in schizonts of Plasmodium falciparum" Parasitology 129(3) 273-287
- ^ "Malaria eModule - SYNCHRONICITY".[dead link]
- ^ Alves E, Bartlett PJ, Garcia CR, Thomas AP (2010). "Melatonin and IP3-induced Ca2+ release from intracellular stores in the malaria parasite Plasmodium falciparum within infected red blood cells. J. Biol". Chem.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ "Malaria eModule - GAMETOCYTOGENESIS".[dead link]
- ^ Sinton JA (1938) The action of Atebrin upon gametocytes of Plasmodium falciparum. Riv Malariol 17:305-330
- ^ Reece SE, Drew DR, Gardner A (2008) Sex ratio adjustment and kin discrimination in malaria parasites" Nature 453(7195) 609-614
- ^ Ponzi M, Siden-Kiamos I, Bertuccinin L, Curra C, Kroeze H, Camarda G, Pace T, Franke-Fayard B, Laurentino EC, Louis K, Waters AP, Janse CJ, Alano P (2009) Egress of Plasmodium berghei gametes from their host erythrocyte is mediated by MDV-1/PEG3 protein. Cell Microbiol 11:1272-1288
- ^ Lal K, Delves MJ, Bromley E, Wastling JM, Tomley FM, Sinden RE (2009) Plasmodium male development gene-1 (mdv-1) is important for female sexual development and identifies a polarised plasma membrane during zygote development. Int J Parasitol 39:755-761
- ^ Robert, V; Read, AF; Essong, J; Tchuinkam, T; Mulder, B; Verhave, JP; Carnevale, P (1996). "Effect of gametocyte sex ratio on infectivity of Plasmodium falciparum to Anopheles gambiae". Trans R Soc Trop Med Hyg. 90 (6): 621–624. doi:10.1016/S0035-9203(96)90408-3. PMID 9015496.
- ^ Teboh-Ewungkem MI, Wang M (2012) Male fecundity and optimal gametocyte sex ratios for Plasmodium falciparum during incomplete fertilization. J Theor Biol
- ^ Sannella AR, Olivieri A, Bertuccini L, Ferre F, Severini C, Pace T, Alano P (2012) Specific tagging of the egress-related osmiophilic bodies in the gametocytes of Plasmodium falciparum. Malar J 11(1) 88
- ^ Tibúrcio M, Silvestrini F, Bertuccini L, Sander A, Turner L, Lavstsen T, Alano P (2012) Early gametocytes of the malaria parasite Plasmodium falciparum specifically remodel the adhesive properties of infected erythrocyte surface. Cell Microbiol doi:10.1111/cmi.12062
- ^ Kangwanrangsan N, Tachibana M, Jenwithisuk R, Tsuboi T, Riengrojpitak S, Torii M, Ishino T (2013) A member of the CPW-WPC protein family is expressed in and localized to the surface of developing ookinetes. Malar J 12(1) 129
- ^ Miao J, Li J, Fan Q, Li X, Li X, Cui L (2010). "The Puf-family RNA-binding protein PfPuf2 regulates sexual development and sex differentiation in the malaria parasite Plasmodium falciparum". J. Cell Sci. 123 (7): 1039. doi:10.1242/jcs.059824.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Laurentino EC, Taylor S, Mair GR, Lasonder E, Bartfai R, Stunnenberg HG, Kroeze H, Ramesar J, Franke-Fayard B, Khan SM, Janse CJ, Waters AP (2011) Experimentally controlled down regulation of the histone chaperone FACT in Plasmodium berghei reveals that it is critical to male gamete fertility. Cell Microbiol doi:10.1111/j.1462-5822.2011.01683.x
- ^ Guttery DS, Ferguson DJ, Poulin B, Xu Z, Straschil U, Klop O, Solyakov L, Sandrini SM, Brady D; et al. (2012). Soldati-Favre, Dominique (ed.). "A Putative Homologue of CDC20/CDH1 in the malaria parasite is essential for male gamete development". PLoS Pathog. 8 (2): e1002554. doi:10.1371/journal.ppat.1002554.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Eksi S, Morahan BJ, Haile Y, Furuya T, Jiang H, Ali O, Xu H, Kiattibutr K, Suri A, Czesny B, Adeyemo A, Myers TG, Sattabongkot J, Su XZ, Williamson KC (2012) Plasmodium falciparum Gametocyte development 1 (Pfgdv1) and gametocytogenesis early gene identification and commitment to sexual development" PLoS Pathog 8(10) e1002964. doi:10.1371/journal.ppat.1002964
- ^ Dvorin, JD; Martyn, DC; Patel, SD; Grimley, JS; Collins, CR; Hopp, CS; Bright, AT; Westenberger, S; Winzeler, E; et al. (2010). "A Plant-Like Kinase in Plasmodium falciparum Regulates Parasite Egress From Erythrocytes". Science. 328 (5980): 910–912. doi:10.1126/science.1188191. PMC 3109083. PMID 20466936.
{{cite journal}}: Explicit use of et al. in:|first9=(help) - ^ Callan-Jones A, Albarran Arriagada OE, Massiera G, Lorman V, Abkarian M (2012) Red blood Cell membrane dynamics during malaria parasite egress. Biophys J. 2012 Dec 19;103(12) 2475-83. doi:10.1016/j.bpj.2012.11.008
- ^ Millholland MG, Chandramohanadas R, Pizarro A, Wehr A, Shi H, Darling C, Lim CT, Greenbaum DC (2011). "The malaria parasite progressively dismantles the host erythrocyte cytoskeleton for efficient egress". Mol Cell Proteomics.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Mantel PY, Hoang AN, Goldowitz I, Potashnikova D, Hamza B, Vorobjev I, Ghiran I, Toner M, Irimia D, Ivanov AR, Barteneva N, Marti M (2013) Malaria-infected erythrocyte-derived microvesicles mediate cellular communication within the parasite population and with the host immune system. Cell Host Microbe 13(5) 521-534 doi:10.1016/j.chom.2013.04.009
- ^ Regev-Rudzki N, Wilson DW, Carvalho TG, Sisquella X, Coleman BM, Rug M, Bursac D, Angrisano F, Gee M, Hill AF, Baum J, Cowman AF (2013) Cell-cell communication between malaria-infected red blood cells via exosome-like vesicles. Cell pii: S0092-8674(13)00504-7. doi:10.1016/j.cell.2013.04.029
- ^ Millholland MG, Mishra S, Dupont CD, Love MS, Patel B, Shilling D, Kazanietz MG, Foskett JK, Hunter CA, Sinnis P, Greenbaum DC (2013) A host GPCR signaling network required for the cytolysis of infected cells facilitates release of apicomplexan parasites. Cell Host Microbe 13(1) 15-28. doi:10.1016/j.chom.2012.12.001
- ^ Dasari P, Reiss K, Lingelbach K, Baumeister S, Lucius R, Udomsangpetch R, Bhakdi SC, Bhakdi S (2011). "Digestive vacuoles of Plasmodium falciparum are selectively phagocytosed by and impair killing function of polymorphonuclear leukocytes". Blood. 118 (18): 4946–56. doi:10.1182/blood-2011-05-353920. PMID 21911835.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Arisue, N; Kawai, S; Hirai, M; Palacpac, NM; Jia, M; Kaneko, A; Tanabe, K; Horii, T (2011). Langsley, Gordon (ed.). "Clues to evolution of the SERA multigene family in 18 Plasmodium species". PLoS ONE. 6 (3): e17775. doi:10.1371/journal.pone.0017775.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b c Ruecker A, Shea M, Hackett F, Suarez C, Hirst EM, Milutinovic K, Withers-Martinez C, Blackman MJ (2012) Proteolytic activation of the essential parasitophorous vacuole cysteine protease SERA6 accompanies malaria parasite egress from its host erythrocyte. J Biol Chem Cite error: The named reference "Ruecker2012" was defined multiple times with different content (see the help page).
- ^ Agarwal S, Singh MK, Garg S, Chitnis CE, Singh S (2012) Ca(2+) Mediated exocytosis of subtilisin-like protease 1: A key step in egress of P. falciparum merozoites. Cell Microbiol doi:10.1111/cmi.12086
- ^ Collins CR, Hackett F, Strath M, Penzo M, Withers-Martinez C, Baker DA, Blackman MJ (2013) Malaria parasite cGMP-dependent protein kinase regulates blood stage merozoite secretory organelle discharge and egress" PLoS Pathog 9(5) e1003344
- ^ Talman AM, Lacroix C, Marques SR, Blagborough AM, Carzaniga R, Ménard R, Sinden RE (2011) PbGEST mediates malaria transmission to both mosquito and vertebrate host. Mol Microbiol doi:10.1111/j.1365-2958.2011.07823.x
- ^ Deligianni E, Morgan RN, Bertuccini L, Wirth CC, Silmon de Monerri NC, Spanos L, Blackman MJ, Louis C, Pradel G, Siden-Kiamos I (2013) A perforin-like protein mediates disruption of the erythrocyte membrane during egress of Plasmodium berghei male gametocytes. Cell Microbiol doi:10.1111/cmi.12131
- ^ Garg S, Agarwal S, Kumar S, Shams Yazdani S, Chitnis CE, Singh S (2013) Calcium-dependent permeabilization of erythrocytes by a perforin-like protein during egress of malaria parasites. Nat. Commun. 4:1736
- ^ Glushakova S, Lizunov V, Blank PS, Melikov K, Humphrey G, Zimmerberg J (2013) Cytoplasmic free Ca2+ is essential for multiple steps in malaria parasite egress from infected erythrocytes. Malar J 12(1) 41
- ^ Engelberg K, Paul AS, Prinz B, Kono M, Ching W, Heincke D, Dobner T, Spielmann T, Duraisingh M, Gilberger TW (2013) Specific phosphorylation of the PfRh2b invasion ligand of Plasmodium falciparum. Biochem J
- ^ Simon N, Lasonder E, Scheuermayer M, Kuehn A, Tews S, Fischer R, Zipfel PF, Skerka C, Pradel G (2012) Malaria parasites co-opt human Factor H to prevent complement-mediated lysis in the mosquito midgut. Cell Host Microbe 13(1) 29-41 doi:10.1016/j.chom.2012.11.013
- ^ Molina-Cruz A, Garver LS, Alabaster A, Bangiolo L, Haile A, Winikor J, Ortega C, van Schaijk BC, Sauerwein RW, Taylor-Salmon E, Barillas-Mury C (2013) The human malaria parasite Pfs47 gene mediates evasion of the mosquito immune system. Science
- ^ Billker, O; Lindo, V; Panico, M; Etienne, AE; Paxton, T; Dell, A; Rogers, M; Sinden, RE; Morris, HR (March 19, 1998). "Identification of xanthurenic acid as the putative inducer of malaria development in the mosquito". Nature. 392 (6673): 289–292. doi:10.1038/32667. PMID 9521324.
{{cite journal}}: Unknown parameter|displayauthors=ignored (|display-authors=suggested) (help) - ^ Eastman RT, Pattaradilokrat S, Raj DK, Dixit S, Deng B, Miura K, Yuan J, Tanaka TQ, Johnson RL, Jiang H, Huang R, Williamson K, Lambert LE, Long C, Austin CP, Wu Y, Su XZ (2012) A class of tricyclic compounds blocking malaria oocyst development and transmission. Antimicrob Agents Chemother
- ^ Portman N, Foster C, Walker G, Slapeta J (2013) Evidence for intraflagellar transport and apical complex formation in a free living relative of the Apicomplexa. Eukaryot Cell
- ^ Wilson LG, Carter LM, Reece SE (2013) High-speed holographic microscopy of malaria parasites reveals ambidextrous flagellar waveforms. Proc Natl Acad Sci USA 2013
- ^ Slavic K, Delves MJ, Prudencio M, Talman AM, Straschil U, Derbyshire ET, Xu Z, Sinden RE, Mota MM, Morin C, Tewari R, Krishna S, Staines HM (2011) Use of a selective inhibitor to define the chemotherapeutic potential of the plasmodial hexose transporter in different stages of the parasite's life cycle" Antimicrob Agents Chemother 55:2824-2830
- ^ a b c Vaughan JA, Noden BH, Beier JC (1992) Population dynamics of Plasmodium falciparum sporogony in laboratory-infected Anopheles gambiae. J Parasitol 78(4) 716-724 Cite error: The named reference "Vaughan1992" was defined multiple times with different content (see the help page).
- ^ Ning J, Otto TD, Pfander C, Schwach F, Brochet M, Bushell E, Goulding D, Sanders M, Lefebvre PA, Pei J, Grishin NV, Vanderlaan G, Billker O, Snell WJ (2013) Comparative genomics in Chlamydomonas and Plasmodium identifies an ancient nuclear envelope protein family essential for sexual reproduction in protists, fungi, plants, and vertebrates" Genes Dev 27(10) 1198-1215
- ^ Li F, Patra KP, Yowell CA, Dame JB, Chin K, Vinetz JM (2010). "Apical surface expression of aspartic protease plasmepsin 4, a potential transmission-blocking target of the Plasmodium ookinete". J. Biol. Chem. 285 (11): 8076–83. doi:10.1074/jbc.M109.063388. PMC 2832958. PMID 20056606.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Hliscs M, Sattler J, Tempel W, Artz JD, Dong A, Hui R, Matuschewski K, Schuler H (2010). "Structure and function of a G-actin sequestering protein with a vital role in malaria oocyst development inside the mosquito vector". J Biol Chem. 285 (15): 11572–83. doi:10.1074/jbc.M109.054916. PMC 2857035. PMID 20083609.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Shimizu, S; Osada, Y; Kanazawa, T; Tanaka, Y; Arai, M (2010). "Suppressive effect of azithromycin on Plasmodium berghei mosquito stage development and apicoplast replication". Malar J. 9 (1): 73. doi:10.1186/1475-2875-9-73.
{{cite journal}}: Unknown parameter|author-separator=ignored (help)CS1 maint: unflagged free DOI (link) - ^ Hernández-Romano J, Rodríguez MH, Pando V, Torres-Monzón JA, Alvarado-Delgado A, Lecona Valera AN, Ramos RA, Martínez-Barnetche J, Rodríguez MC (2011). "Conserved peptide sequences bind to actin and enolase on the surface of Plasmodium berghei ookinetes". Parasitology. 138 (11): 1341–53. doi:10.1017/S0031182011001296. PMID 21816124.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Siden-Kiamos I, Ganter M, Kunze A, Hliscs M, Steinbüchel M, Mendoza J, Sinden RE, Louis C, Matuschewski K (2011) Stage-specific depletion of Myosin A supports an essential role in motility of malarial ookinetes. Cell Microbiol doi:10.1111/j.1462-5822.2011.01686.x
- ^ a b Aly AS, Matuschewski K (2005). "A malarial cysteine protease is necessary for Plasmodium sporozoite egress from oocysts". J Exp Med. 202 (2): 225–230. doi:10.1084/jem.20050545. PMC 2213010. PMID 16027235. Cite error: The named reference "Aly2005" was defined multiple times with different content (see the help page).
- ^ Philip N, Vaikkinen HJ, Tetley L, Waters AP (2012) A unique kelch domain phosphatase in Plasmodium regulates ookinete morphology, motility and invasion" PLoS One 7(9) e44617
- ^ Kutuzov MA, Andreeva AV (2002) Protein Ser/Thr phosphatases with kelch-like repeat domains. Cell Signal 14: 745–750
- ^ Reininger L, Billker O, Tewari R, Mukhopadhyay A, Fennell C, et al. (2005) A NIMA-related protein kinase is essential for completion of the sexual cycle of malaria parasites" J Biol Chem 280: 31957–31964
- ^ a b c Reininger L, Tewari R, Fennell C, Holland Z, Goldring D, et al. (2009) An essential role for the Plasmodium Nek-2 Nima-related protein kinase in the sexual development of malaria parasites" J Biol Chem 284: 20858–20868 Cite error: The named reference "Reininger2009" was defined multiple times with different content (see the help page).
- ^ Ellekvist P, Maciel J, Mlambo G, Ricke CH, Colding H, Klaerke DA, Kumar N (2008) Critical role of a K+ channel in Plasmodium berghei transmission revealed by targeted gene disruption" Proc Natl Acad Sci USA 105(17) 6398-6402
- ^ Mlambo G, Coppens I, Kumar N (2012) Aberrant sporogonic development of Dmc1 (a meiotic recombinase) deficient Plasmodium berghei parasites" PLoS One 7(12) e52480. doi:10.1371/journal.pone.0052480
- ^ Zollner GE, Ponsa N, Garman GW, Poudel S, Bell JA, Sattabongkot J, Coleman RE, Vaughan JA (2006) Population dynamics of sporogony for Plasmodium vivax parasites from western Thailand developing within three species of colonized Anopheles mosquitoes. Malar J 5:68
- ^ Kudryashev M, Lepper S, Stanway R; et al. (2010). "Positioning of large organelles by a membrane- associated cytoskeleton in Plasmodium sporozoites". Cell. Microbiol. 12 (3): 362–71. doi:10.1111/j.1462-5822.2009.01399.x. PMID 19863555.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Moraes CB, Dorval T, Contreras-Dominguez M, Dossin Fde M, Hansen MA, Genovesio A, Freitas-Junior LH (2013) Transcription sites are developmentally regulated during the asexual cycle of Plasmodium falciparum. 8(2) e55539. doi:10.1371/journal.pone.0055539
- ^ Tremp AZ, Khater EI, Dessens JT (2008) IMC1b is a putative membrane skeleton protein involved in cell shape, mechanical strength, motility, and infectivity of malaria ookinetes" J Biol Chem 283(41) 27604-27611
- ^ Tremp A, Carter V, Saeed S, Dessens JT (2013) Morphogenesis of Plasmodium zoites is uncoupled from tensile strength. Mol Microbiol doi:10.1111/mmi.12297
- ^ Zuccala ES, Gout AM, Dekiwadia C, Marapana DS, Angrisano F, Turnbull L, Riglar DT, Rogers KL, Whitchurch CB, Ralph SA, Speed TP, Baum J (2012) Subcompartmentalisation of proteins in the rhoptries correlates with ordered events of erythrocyte invasion by the blood stage malaria parasite" PLoS One 7(9) e46160
- ^ Kemp LE, Yamamoto M, Soldati-Favre D (2012) Subversion of host cellular functions by the apicomplexan parasites. FEMS Microbiol Rev doi:10.1111/1574-6976.12013
- ^ Straub KW, Peng ED, Hajagos BE, Tyler JS, Bradley PJ (2011). Striepen, Boris (ed.). "The moving junction protein RON8 facilitates firm attachment and host cell invasion in Toxoplasma gondii". PLoS Pathog. 7 (3): e1002007. doi:10.1371/journal.ppat.1002007.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Moreno-Perez DA, Montenegro M, Patarroyo ME, Patarroyo MA (2011). "Identification, characterization and antigenicity of the Plasmodium vivax rhoptry neck protein 1 (PvRON1)". Malar. J. 10: 314. doi:10.1186/1475-2875-10-314. PMC 3215230. PMID 22024312.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Wang B, Lu F, Cheng Y, Li J, Ito D, Sattabongkot J, Tsuboi T, Han ET (2012) Identification and characterization of the Plasmodium falciparum RhopH2 ortholog in Plasmodium vivax. Parasitol Res
- ^ Srinivasan P, Beatty WL, Diouf A, Herrera R, Ambroggio X, Moch JK, Tyler JS, Narum DL, Pierce SK, Boothroyd JC, Haynes JD, Miller LH (2011). "Binding of Plasmodium merozoite proteins RON2 and AMA1 triggers commitment to invasion". Proc Natl Acad Sci U S A. 108 (32): 13275–80. doi:10.1073/pnas.1110303108. PMC 3156155. PMID 21788485.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Tang J, Dai Y, Zhang H, Culleton RL, Liu Y, Zhao S, Wang X, Guan X, Kaneko O, Zhu Y (2012) Positive diversifying selection on Plasmodium vivax RON2 protein. Parasitology
- ^ Ghoneim AM (2013) Trafficking of Plasmodium falciparum chimeric rhoptry protein with Brefeldin A. Folia Parasitol (Praha) 60(1) 75-78
- ^ Cabrera A, Herrmann S, Warszta D, Santos JM, John Peter AT, Kono M, Debrouver S, Jacobs T, Spielmann T, Ungermann C, Soldati-Favre D, Gilberger TW (2012) Dissection of minimal sequence requirements for rhoptry membrane targeting in the malaria parasite" Traffic 9999(999A). doi:10.1111/j.1600-0854.2012.01394.x
- ^ Frénal K, Tay CL, Mueller C, Bushell ES, Jia Y, Graindorge A, Billker O, Rayner JC, Soldati-Favre D (2013) Global analysis of apicomplexan protein S-acyl transferases reveals an enzyme essential for invasion. Traffic doi:10.1111/tra.12081
- ^ Olivieri A, Collins CR, Hackett F, Withers-Martinez C, Marshall J, Flynn HR, Skehel JM, Blackman MJ (2011). Carruthers, Vern B (ed.). "Juxtamembrane shedding of Plasmodium falciparum AMA1 is sequence independent and essential, and helps evade invasion-inhibitory antibodies". PLoS Pathog. 7 (12): e1002448. doi:10.1371/journal.ppat.1002448.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Bargieri DY, Andenmatten N, Lagal V, Thiberge S, Whitelaw JA, Tardieux I, Meissner M, Ménard R (2013) Apical membrane antigen 1 mediates apicomplexan parasite attachment but is dispensable for host cell invasion. Nat Commun 4:2552. doi:10.1038/ncomms3552
- ^ Vulliez-Le Normand, B; Tonkin, ML; Lamarque, MH; Langer, S; Hoos, S; Roques, M; Saul, FA; Faber, BW; Bentley, GA; Boulanger, Martin J.; Lebrun, Maryse; et al. (2012). Phillips, Meg (ed.). "Structural and functional insights into the malaria parasite moving junction complex". PLoS Pathog. 8 (6): e1002755. doi:10.1371/journal.ppat.1002755.
{{cite journal}}: Explicit use of et al. in:|first9=(help); Unknown parameter|month=ignored (help)CS1 maint: unflagged free DOI (link) - ^ Hegge S, Münter S, Steinbüchel M, Heiss K, Engel U, Matuschewski K, Frischknecht F (2010). "Multistep adhesion of Plasmodium sporozoites". FASEB J. 24 (7): 2222–34. doi:10.1096/fj.09-148700. PMID 20159960.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Wickert H, Göttler W, Krohne G, Lanzer M (2004) Maurer's cleft organization in the cytoplasm of Plasmodium falciparum-infected erythrocytes: new insights from three-dimensional reconstruction of serial ultrathin sections. Eur J Cell Biol 83(10) 567-582
- ^ Tsarukyanova I, Drazba JA, Fujioka H, Yadav SP, Sam-Yellowe TY (2009). "Proteins of the Plasmodium falciparum two transmembrane Maurer's cleft protein family, PfMC-2TM, and the 130 kDa Maurer's cleft protein define different domains of the infected erythrocyte intramembranous network". Parasitol Res. 104 (4): 875–891. doi:10.1007/s00436-008-1270-3. PMID 19130087.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Spycher C, Rug M, Klonis N, Ferguson DJ, Cowman AF, Beck HP, Tilley L (2006). "Genesis of and Trafficking to the Maurer's Clefts of Plasmodium falciparum-Infected Erythrocytes". Mol Cell Biol. 26 (11): 4074–4085. doi:10.1128/MCB.00095-06. PMC 1489082. PMID 16705161.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Cooke BM, Buckingham DW, Glenister FK, Fernandez KM, Bannister LH, Marti M, Mohandas N, Coppel RL (2006). "A Maurer's cleft–associated protein is essential for expression of the major malaria virulence antigen on the surface of infected red blood cells". J Cell Biol. 172 (6): 899–908. doi:10.1083/jcb.200509122. PMC 2063733. PMID 16520384.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Spycher C, Klonis N, Spielmann T, Kump E, Steiger S, et al (2003) MAHRP-1, a novel Plasmodium falciparum histidine-rich protein, binds ferriprotoporphyrin IX and localizes to the Maurer's clefts" J Biol Chem 278: 35373–35383
- ^ Hawthorne PL, Trenholme KR, Skinner-Adams TS, Spielmann T, Fischer K, et al A novel Plasmodium falciparum ring stage protein, REX, is located in Maurer's clefts. Mol Biochem Parasitol 136: 181–189
- ^ Dixon MW, Kenny S, McMillan PJ, Hanssen E, Trenholme KR, Gardiner DL, Tilley L (2011) Genetic ablation of a Maurer's cleft protein prevents assembly of the Plasmodium falciparum virulence complex. Mol Microbiol doi:10.1111/j.1365-2958.2011.07740.x
- ^ Atkinson CT, Aikawa M, Perry G, Fujino T, Bennett V, Davidson EA, Howard RJ (1988) Ultrastructural localization of erythrocyte cytoskeletal and integral membrane proteins in Plasmodium falciparum-infected erythrocytes. Eur J Cell Biol 45(2) 192-199
- ^ a b Cyrklaff M, Sanchez CP, Kilian N, Bisseye C, Simpore J, Frischknecht F, Lanzer M (2012) Hemoglobins S and C interfere with actin remodeling in Plasmodium falciparum-infected erythrocytes" Science 334(6060) 1283-1286 Cite error: The named reference "Cyrklaff2012" was defined multiple times with different content (see the help page).
- ^ Kulangara C, Luedin S, Dietz O, Rusch S, Frank G, Mueller D, Moser M, Kajava AV, Corradin G, Beck HP, Felger I (2012) Cell biological characterization of the malaria vaccine candidate trophozoite exported protein 1" PLoS One 7(10) e46112. doi:10.1371/journal.pone.0046112
- ^ Hanssen E, Hawthorne P, Dixon MW, Trenholme KR, McMillan PJ et al (2008) Targeted mutagenesis of the ring-exported protein-1 of Plasmodium falciparum disrupts the architecture of Maurer's cleft organelles" Mol Microbiol 69: 938–953
- ^ a b Glenister FK, Fernandez KM, Kats LM, Hanssen E, Mohandas N et al (2009) Functional alteration of red blood cells by a megadalton protein of Plasmodium falciparum" Blood 113: 919–928
- ^ Kilian N, Dittmer M, Cyrklaff M, Ouermi D, Bisseye C, Simpore J, Frischknecht F, Sanchez CP, Lanzer M (2012) Hemoglobin S and C affect the motion of Maurer's clefts in P. falciparum-infected erythrocytes. Cell Microbiol doi:10.1111/cmi.12102
- ^ a b Zhu X, Yahata K, Alexandre JS, Tsuboi T, Kaneko O (2012) The N-terminal segment of Plasmodium falciparum SURFIN(4.1) is required for its trafficking to the red blood cell cytosol through the endoplasmic reticulum. Parasitol Int pii: S1383-5769(12)00166-3. doi:10.1016/j.parint.2012.12.006
- ^ McMillan PJ, Millet C, Batinovic S, Maiorca M, Hanssen E, Kenny S, Muhle RA, Melcher M, Fidock DA, Smith JD, Dixon MW, Tilley L (2013) Spatial and temporal mapping of the PfEMP1 export pathway in Plasmodium falciparum. Cell Microbiol doi:10.1111/cmi.12125
- ^ Mbengue A, Audiger N, Vialla E, Dubremetz JF, Braun-Breton C (2013) Novel Plasmodium falciparum Maurer's clefts protein families implicated in the release of infectious merozoites. Mol Microbiol doi:10.1111/mmi.12193
- ^ Frech C, Chen N (2013) Variant surface antigens of malaria parasites: functional and evolutionary insights from comparative gene family classification and analysis" BMC Genomics 14(1) 427
- ^ Hayton K, Templeton TJ (2008) Osmiophilic bodies and the odd organelles of alveolates" Mol Microbiol 67(2) 236-240
- ^ Trager W, Rozario C, Shio H, Williams J, Perkins ME (1992) Transfer of a dense granule protein of Plasmodium falciparum to the membrane of ring stages and isolation of dense granules" Infect Immun 60(11) 4656-4661
- ^ Abu Bakar N, Klonis N, Hanssen E, Chan C, Tilley L (2010). "Digestive-vacuole genesis and endocytic processes in the early intraerythrocytic stages of Plasmodium falciparum". J Cell Sci.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Vaid A, Ranjan R, Smythe WA, Hoppe HC, Sharma P (2010). "PfPI3K, a Phosphatidylinositol-3 kinase from Plasmodium falciparum, is exported to the host erythrocyte and is involved in hemoglobin trafficking". Blood. 115 (12): 2500–7. doi:10.1182/blood-2009-08-238972. PMC 2918364. PMID 20093402.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Eksi S, Williamson KC (2011). "Protein targeting to the parasitophorous vacuole membrane of Plasmodium falciparum.". Eukaryotic Cell. 10 (6): 744. doi:10.1128/EC.00008-11.
- ^ Vera IM, Beatty WL, Sinnis P, Kim K (2011). Deitsch, Kirk (ed.). "Plasmodium protease ROM1 is important for proper formation of the parasitophorous vacuole". PLoS Pathog. 7 (9): e1002197. doi:10.1371/journal.ppat.1002197.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ van de Hoef DL, Coppens I, Holowka T, Ben Mamoun C, Branch O, Rodriguez A (2013) Plasmodium falciparum-derived uric acid precipitates induce maturation of dendritic cells" PLoS One 8(2) e55584. doi:10.1371/journal.pone.0055584
- ^ van Ooij C, Withers-Martinez C, Ringel A, Cockcroft S, Haldar K, Blackman MJ (2013) Identification of a Plasmodium falciparum phospholipid transfer protein. J Biol Chem
- ^ Biot C, Botté CY, Dubar F, Maréchal E (2002) Targeting malaria parasite at the level of apicoplast: an update. Med Sci (Paris) 28(2) 163-171. doi:10.1051/medsci/2012282014
- ^ Lemgruber L, Kudryashev M, Dekiwadia C, Riglar DT, Baum J, Stahlberg H, Ralph SA, Frischknecht F (2013) Cryo-electron tomography reveals four-membrane architecture of the Plasmodium apicoplast. Malar J 12(1) 25
- ^ Arisue N, Hashimoto T, Mitsui H, Palacpac NM, Kaneko A, Kawai S, Hasegawa M, Tanabe K, Horii T (2012) The Plasmodium apicoplast genome: conserved structure and close relationship of P. ovale to rodent malaria parasites. Mol Biol Evol
- ^ Kumar A, Tanveer A, Biswas S, Ram ER, Gupta A, Kumar B, Habib S (2010). "Nuclear-encoded DnaJ homolog of Plasmodium falciparum interacts with replication ori of the apicoplast genome". Mol. Microbiol. 75 (4): 942–56. doi:10.1111/j.1365-2958.2009.07033.x. PMID 20487289.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Kennedy SR, Chen CY, Schmitt MW, Bower CN, Loeb LA (2011). "The biochemistry and fidelity of synthesis by the apicoplast genome replication DNA polymerase Pfprex from the malaria parasite Plasmodium falciparum". J Mol Biol. 410 (1): 27–38. doi:10.1016/j.jmb.2011.04.071. PMC 3117635. PMID 21570407.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Gallagher JR, Matthews KA, Prigge ST (2011) P. falciparum apicoplast transit peptides are unstructured in vitro and during apicoplast import. Traffic doi:10.1111/j.1600-0854.2011.01232.x
- ^ Tonkin CJ, Roos DS, McFadden GI (2006) N-terminal positively charged amino acids, but not their exact position, are important for apicoplast transit peptide fidelity in Toxoplasma gondii. Mol Biochem Parasitol 150(2) 192-200
- ^ Tawk L, Dubremetz JF, Montcourrier P, Chicanne G, Merezegue F, Richard V, Payrastre B, Meissner M, Vial HJ, Roy C, Wengelnik K, Lebrun M (2011) Phosphatidylinositol 3-monophosphate is involved in toxoplasma apicoplast biogenesis. PloS Pathog. 7(2) e1001286
- ^ Biot C, Botté CY, Dubar F, Maréchal E (2012) Targeting malaria parasite at the level of apicoplast: an update. Med Sci (Paris) 28(2) 163-171
- ^ a b Deschermeier C, Hecht LS, Bach F, Rützel K, Stanway RR, Nagel A, Seeber F, Heussler VT (2011) Mitochondrial lipoic acid scavenging is essential for Plasmodium berghei liver stage development. Cell Microbiol doi:10.1111/j.1462-5822.2011.01729.x Cite error: The named reference "Deschermeier2011" was defined multiple times with different content (see the help page).
- ^ Yeh E, Derisi JL (2011). Striepen, Boris (ed.). "Chemical rescue of malaria parasites lacking an apicoplast defines organelle function in blood-stage Plasmodium falciparum". PLoS Biol. 9 (8): e1001138. doi:10.1371/journal.pbio.1001138.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Kumar B, Chaubey S, Shah P, Tanveer A, Charan M, Siddiqi MI, Habib S (2011). "Interaction between sulphur mobilisation proteins SufB and SufC: Evidence for an iron-sulphur cluster biogenesis pathway in the apicoplast of Plasmodium falciparum". Int J Parasitol. 41 (9): 991–9. doi:10.1016/j.ijpara.2011.05.006. PMID 21722645.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Seeber F, Soldati-Favre D (2010) Metabolic pathways in the apicoplast of apicomplexa. Int Rev Cell Mol Biol 281: 161–228
- ^ Gisselberg JE, Dellibovi-Ragheb TA, Matthews KA, Bosch G, Prigge ST (2013) The suf iron-sulfur cluster synthesis pathway is required for apicoplast maintenance in malaria parasites" PLoS Pathog 9(9) e1003655
- ^ Haussig JM, Matuschewski K, Kooij TW (2013) Experimental genetics of Plasmodium berghei NFU in the apicoplast iron-sulfur cluster biogenesis pathway" PLoS One 8(6) e67269
- ^ Haussig JM, Matuschewski K, Kooij TW (2011) Inactivation of a Plasmodium apicoplast protein attenuates formation of liver merozoites. Mol Microbiol doi:10.1111/j.1365-2958.2011.07787.x
- ^ a b Ponpuak M, Klemba M, Park M, Gluzman IY, Lamppa GK, Goldberg DE (2006). "A role for falcilysin in transit peptide degradation in the Plasmodium falciparum apicoplast". Mol Microbiol. 63 (2): 314–334. doi:10.1111/j.1365-2958.2006.05443.x. PMID 17074076.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Chaudhari R, Narayan A, Patankar S (2012) A novel trafficking pathway in Plasmodium falciparum for the organellar localization of glutathione peroxidase-like thioredoxin peroxidase. FEBS J doi:10.1111/j.1742-4658.2012.08746.x
- ^ Kitamura K, Kishi-Itakura C, Tsuboi T, Sato S, Kita K, Ohta N, Mizushima N (2012) Autophagy-related Atg8 localizes to the apicoplast of the human malaria parasite Plasmodium falciparum" PLoS One 7(8) e42977.
- ^ Banerjee T, Jaijyan DK, Surolia N, Singh AP, Surolia A (2012) Apicoplast triose phosphate transporter (TPT) gene knockout is lethal for Plasmodium. Mol Biochem Parasitol pii: S0166-6851(12)00241-1. doi:10.1016/j.molbiopara.2012.09.008
- ^ Foth BJ, Stimmler LM, Handman E, Crabb BS, Hodder AN, McFadden GI (2005) The malaria parasite Plasmodium falciparum has only one pyruvate dehydrogenase complex, which is located in the apicoplast" Mol Microbiol 55(1) 39-53
- ^ a b Cobbold SA, Vaughan AM, Lewis IA, Painter HJ, Camargo N, Perlman DH, Fishbaugher M, Healer J, Cowman AF, Kappe SH, Llinás M (2013) Kinetic flux profiling elucidates two independent acetyl-CoA biosynthetic pathways in Plasmodium falciparum. J Biol Chem
- ^ El Bakkouri M, Rathore S, Calmettes C, Wernimont AK, Liu K, Sinha D, Asad M, Jung P, Hui R, Mohmmed A, Houry WA (2012) Structural insights into the inactive subunit of the apicoplast-localized caseinolytic protease complex of Plasmodium falciparum. J Biol Chem
- ^ Botté CY, Yamaryo-Botté Y, Rupasinghe TW, Mullin KA, Macrae JI, Spurck TP, Kalanon M, Shears MJ, Coppel RL, Crellin PK, Maréchal E, McConville MJ, McFadden GI (2013) Atypical lipid composition in the purified relict plastid (apicoplast) of malaria parasites. Proc Natl Acad Sci USA
- ^ Smith MH, Ploegh HL, Weissman JS (2011) Road to ruin: targeting proteins for degradation in the endoplasmic reticulum" Science 334: 1086–1090. doi:10.1126/science.1209235
- ^ Ngansop F, Li H, Zolkiewska A, Zolkiewski M (2013) Biochemical characterization of the apicoplast-targeted AAA+ ATPase ClpB from Plasmodium falciparum. Biochem Biophys Res Commun pii: S0006-291X(13)01411-3. doi:10.1016/j.bbrc.2013.08.064
- ^ Mailu BM, Ramasamy G, Mudeppa DG, Li L, Lindner SE, Peterson MJ, Derocher AE, Kappe SH, Rathod PK, Gardner MJ (2013) A non-discriminating glutamyl-tRNA synthetase in the Plasmodium apicoplast: The first enzyme in an indirect aminoacylation pathway. J Biol Chem
- ^ a b c Gupta A, Mir SS, Saqib U, Biswas S, Vaishya S, Srivastava K, Siddiqi MI, Habib S (2013) The effect of fusidic acid on Plasmodium falciparum elongation factor G (EF-G). Mol Biochem Parasitol. 2013 Nov 6. pii: S0166-6851(13)00151-5. doi: 10.1016/j.molbiopara.2013.10.003 Cite error: The named reference "Gupta2013" was defined multiple times with different content (see the help page).
- ^ McMillan PJ, Stimmler LM, Foth BJ, McFadden GI, Müller S (2005). "The human malaria parasite Plasmodium falciparum possesses two distinct dihydrolipoamide dehydrogenases". Molecular Microbiology. 55 (1): 27–38. doi:10.1111/j.1365-2958.2004.04398.x. PMID 15612914.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Foth BJ, Stimmler LM, Handman E, Crabb BS, Hodder AN, McFadden GI (2005). "The malaria parasite Plasmodium falciparum has only one pyruvate dehydrogenase complex, which is located in the apicoplast". Molecular Microbiology. 55 (1): 39–53. doi:10.1111/j.1365-2958.2004.04407.x. PMID 15612915.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Balabaskaran Nina P, Morrisey JM, Ganesan SM, Ke H, Pershing AM, Mather MW, Vaidya AB (2011). "ATP synthase complex of Plasmodium falciparum: dimeric assembly in mitochondrial membranes and resistance to genetic disruption". J Biol Chem.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Eckers E, Petrungaro C, Gross D, Riemer J, Hell K, Deponte M (2012) Divergent molecular evolution of the mitochondrial sulfhydryl:cytochrome c oxidoreductase Erv in opisthokonts and parasitic protists. J Biol Chem
- ^ Hino A, Hirai M, Tanaka TQ, Watanabe YI, Matsuoka H, Kita K (2012) Critical roles of the mitochondrial complex II in oocyst formation of rodent malaria parasite Plasmodium berghei. J Biochem
- ^ Tanaka TQ, Hirai M, Watanabe YI, Kita K (2012) Towards understanding the role of mitochondrial complex II in the intraerythrocytic stages of Plasmodium falciparum: Gene targeting of the Fp subunit. Parasitol Int
- ^ Nozawa A, Fujimoto R, Matsuoka H, Tsuboi T, Tozawa Y (2011) Biochem Biophys Res Commun
- ^ Rathore S, Jain S, Sinha D, Gupta M, Asad M, Srivastava A, Narayanan MS, Ramasamy G, Chauhan VS; et al. (2011). "Disruption of a mitochondrial protease machinery in Plasmodium falciparum is an intrinsic signal for parasite cell death". Cell Death Dis. 2 (11): e231. doi:10.1038/cddis.2011.118.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Fisher N, Abd Majid R, Antoine T, Al-Helal M, Warman AJ, Johnson DJ, Lawrenson AS, Ranson H, O'Neill PM, Ward SA, Biagini GA (2012). "Cytochrome b mutation Y268S conferring the atovaquone resistance phenotype in the malaria parasite results in reduced parasite bc1 catalytic turnover and protein expression". J Biol Chem. 287 (13): 9731–41. doi:10.1074/jbc.M111.324319. PMC 3322985. PMID 22282497.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Ke H, Morrisey J, Ganesan SM, Mather MW, Vaidya AB (2012) Mitochondrial RNA polymerase is an essential enzyme in erythrocytic stages of Plasmodium falciparum. Mol Biochem Parasitol
- ^ Feagin JE, Harrell MI, Lee JC, Coe KJ, Sands BH, Cannone JJ, Tami G, Schnare MN, Gutell RR (2012) The fragmented mitochondrial ribosomal RNAs of Plasmodium falciparum" PLoS One 7(6) e38320.
- ^ Masuda-Suganuma H, Usui M, Fukumoto S, Inoue N, Kawazu SI (2012) Mitochondrial peroxidase TPx-2 is not essential in the blood and insect stages of Plasmodium berghei. Parasit Vectors 5(1) 252
- ^ Jain S, Rathore S, Asad M, Hossain ME, Sinha D, Datta G, Mohmmed A (2013) The prokaryotic ClpQ protease plays a key role in growth and development of mitochondria in Plasmodium falciparum. Cell Microbiol doi:10.1111/cmi.12142
- ^ Vallières C, Fisher N, Meunier B (2013) Reconstructing the Qo Site of Plasmodium falciparum bc 1 complex in the yeast enzyme" PLoS One 8(8) e71726. doi:10.1371/journal.pone.0071726
- ^ Tanveer A, Allen SM, Jackson KE, Charan M, Ralph SA, Habib S (2013) An FtsH protease is recruited to the mitochondrion of Plasmodium falciparum" PLoS One 8(9) e74408. doi:10.1371/journal.pone.0074408
- ^ Wunderlich J, Rohrbach P, Dalton JP (2012) The malaria digestive vacuole. Front Biosci (Schol Ed) 4:1424-1448
- ^ Ch'ng JH, Liew K, Goh AS, Sidhartha E, Tan KS (2011). "Drug-induced permeabilization of parasite's digestive vacuole is a key trigger of programmed cell death in Plasmodium falciparum". Cell Death Dis. 2 (10): e216. doi:10.1038/cddis.2011.97.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Bohórquez EB, Chua M, Meshnick SR (2012) Quinine localizes to a non-acidic compartment within the food vacuole of the malaria parasite Plasmodium falciparum. Malar J
- ^ a b Dasari P, Heber SD, Beisele M, Torzewski M, Reifenberg K, Orning C, Fries A, Zapf AL, Baumeister S, Lingelbach K, Udomsangpetch R, Bhakdi SC, Reiss K, Bhakdi S (2012) Digestive vacuole of Plasmodium falciparum released during erythrocyte rupture dually activates complement and coagulation. Blood Cite error: The named reference "Dasari2012" was defined multiple times with different content (see the help page).
- ^ a b Ehlgen, F; Pham, JS; De Koning-Ward, T; Cowman, AF; Ralph, SA (2012). Snounou, Georges (ed.). "Investigation of the Plasmodium falciparum food vacuole through inducible expression of the chloroquine resistance transporter (PfCRT)". PLoS ONE. 7 (6): e38781. doi:10.1371/journal.pone.0038781.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Griffin CE, Hoke JM, Samarakoon U, Duan J, Mu J, Ferdig MT, Warhurst DC, Cooper RA (2012) Mutation in the Plasmodium falciparum CRT protein determines the stereospecific activity of the antimalarial Cinchona alkaloids. Antimicrob Agents Chemother
- ^ a b c Kapishnikov S, Weiner A, Shimoni E, Guttmann P, Schneider G, Dahan-Pasternak N, Dzikowski R, Leiserowitz L, Elbaum M (2012) Oriented nucleation of hemozoin at the digestive vacuole membrane in Plasmodium falciparum. Proc Natl Acad Sci USA Cite error: The named reference "Kapishnikov2012" was defined multiple times with different content (see the help page).
- ^ Orjih AU, Mathew TC, Cherian PT (2012) Erythrocyte membranes convert monomeric ferriprotoporphyrin IX to β-hematin in acidic environment at malarial fever temperature. Exp Biol Med (Maywood)
- ^ Prasad R, Atul, Kolla VK, Legac J, Singhal N, Navale R, Rosenthal PJ, Sijwali PS (2013) Blocking Plasmodium falciparum development via dual inhibition of hemoglobin degradation and the ubiquitin proteasome system by MG132" PLoS One 8(9) e73530. doi:10.1371/journal.pone.0073530
- ^ Guizetti J, Martins RM, Guadagnini S, Claes A, Scherf A (2013) Nuclear pores and perinuclear expression sites of var and rDNA genes correspond to physically distinct regions in Plasmodium falciparum. Eukaryotic Cell
- ^ Dahan-Pasternak N, Nasereddin A, Kolevzon N, Pe'er M, Wong W, Shinder V, Turnbull L, Whitchurch CB, Elbaum M, Gilberger TW, Yavin E, Baum J, Dzikowski R (2013) PfSec13 is an unusual chromatin associated nucleoporin of Plasmodium falciparum, which is essential for parasite proliferation in human erythrocytes. J Cell Sci
- ^ a b c Figueiredo LM, Rocha EP, Mancio-Silva L, Prevost C, Hernandez-Verdun D, Scherf A (2005). "The unusually large Plasmodium telomerase reverse-transcriptase localizes in a discrete compartment associated with the nucleolus". Nucleic Acids Res. 33 (3): 1111–1122. doi:10.1093/nar/gki260. PMC 549419. PMID 15722485.
{{cite journal}}: CS1 maint: multiple names: authors list (link) Cite error: The named reference "Figueiredo2005" was defined multiple times with different content (see the help page). - ^ Mancio-Silva L, Lopez-Rubio JJ, Claes A, Scherf A (2013) Sir2a regulates rDNA transcription and multiplication rate in the human malaria parasite Plasmodium falciparum. Nat Commun 4:1530. doi:10.1038/ncomms2539
- ^ Mancio-Silva L, Zhang Q, Scheidig-Benatar C, Scherf A (2010) Clustering of dispersed ribosomal DNA and its role in gene regulation and chromosome-end associations in malaria parasites" Proc Natl Acad Sci USA 107(34) 15117-15122. doi:10.1073/pnas.1001045107
- ^ a b Chung DW, Ponts N, Prudhomme J, Rodrigues EM, Le Roch KG (2012) Characterization of the ubiquitylating components of the human malaria parasite's protein degradation pathway" PLoS One 7(8) e43477
- ^ Huang F, Tang L, Yang H, Zhou S, Liu H, Li J, Guo S (2012) Molecular epidemiology of drug resistance markers of Plasmodium falciparum in Yunnan Province, China. Malar J 11:243
- ^ Phompradit P, Wisedpanichkij R, Muhamad P, Chaijaroenkul W, Na-Bangchang K (2012) Molecular analysis of pfatp6 and pfmdr1 polymorphisms and their association with in vitro sensitivity in Plasmodium falciparum isolates from the Thai-Myanmar border. Acta Trop. 2011 Oct-Nov;120(1-2) 130-5. doi:10.1016/j.actatropica.2011.07.003
- ^ Tahar R, Ringwald P, Basco LK (2009) Molecular epidemiology of malaria in Cameroon. XXVIII. In vitro activity of dihydroartemisinin against clinical isolates of Plasmodium falciparum and sequence analysis of the P. falciparum ATPase 6 gene" Am J Trop Med Hyg 81(1) 13-18
- ^ Pulcini S, Staines HM, Pittman JK, Slavic K, Doerig C, Halbert J, Tewari R, Shah F, Avery MA, Haynes RK, Krishna S (2013) Expression in yeast links field polymorphisms in PfATP6 to in vitro artemisinin resistance and identifies new inhibitor classes. J Infect Dis
- ^ Bhattacharjee S, Stahelin RV, Speicher KD, Speicher DW, Haldar K (2012). "Endoplasmic reticulum PI(3)P lipid binding targets malaria proteins to the Host Cell". Cell. 148 (1–2): 201–212. doi:10.1016/j.cell.2011.10.051. PMC 3268671. PMID 22265412.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Nishimoto, Y; Arisue, N; Kawai, S; Escalante, AA; Horii, T; Tanabe, K; Hashimoto, T. (2008). "Evolution and phylogeny of the heterogeneous cytosolic SSU rRNA genes in the genus Plasmodium". Mol Phylogenet Evol. 47 (1): 45–53. doi:10.1016/j.ympev.2008.01.031. PMID 18334303.
{{cite journal}}: Unknown parameter|author-separator=ignored (help) - ^ a b c d Das S, Basu H, Korde R, Tewari R, Sharma S (2012) Arrest of nuclear division in Plasmodium through blockage of erythrocyte surface exposed ribosomal protein P2" PLoS Pathog 8(8) e1002858 Cite error: The named reference "Das2012" was defined multiple times with different content (see the help page).
- ^ Rohloff P, Miranda K, Rodrigues JC, Fang J, Galizzi M, Plattner H, Hentschel J, Moreno SN (2011) Calcium uptake and proton transport by acidocalcisomes of Toxoplasma gondii" PLoS One 6(4) e18390. doi:10.1371/journal.pone.0018390
- ^ Hellmann JK; Münter S; Kudryashev M; Schulz S; Heiss K; Müller AK; Matuschewski K; Spatz JP; Schwarz US (2011). Mota, Maria M (ed.). "Environmental constraints guide migration of malaria parasites during transmission". PLoS Pathog. 7 (6): e1002080. doi:10.1371/journal.ppat.1002080.
{{cite journal}}:|first10=missing|last10=(help); Invalid|display-authors=9(help); Unknown parameter|author-separator=ignored (help)CS1 maint: unflagged free DOI (link) - ^ Song G, Koksal AC, Lu C, Springer TA (2012) Shape change in the receptor for gliding motility in Plasmodium sporozoites. Proc Natl Acad Sci USA
- ^ Pihlajamaa T, Kajander T, Knuuti J, Horkka K, Sharma A, Permi P (2013) Structure of Plasmodium falciparum thrombospondin-related anonymous protein (TRAP) A domain highlights distinct features in apicomplexan von Willebrand Factor A homologues. Biochem J
- ^ a b Ejigiri I, Ragheb DR, Pino P, Coppi A, Bennett BL, Soldati-Favre D, Sinnis P (2012) Shedding of TRAP by a rhomboid protease from the malaria sporozoite surface is essential for gliding motility and sporozoite infectivity" PLoS Pathog 8(7) e1002725
- ^ Ramakrishnan C, Dessens JT, Armson R, Pinto SB, Talman AM, Blagborough AM, Sinden RE (2011). "Vital functions of the malarial ookinete protein, CTRP, reside in the A domains". Int J Parasitol. 41 (10): 1029–39. doi:10.1016/j.ijpara.2011.05.007. PMID 21729699.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Bergmann-Leitner ES, Legler PM, Savranskaya T, Ockenhouse CF, Angov E (2011). "Cellular and humoral immune effector mechanisms required for sterile protection against sporozoite challenge induced with the novel malaria vaccine candidate CelTOS". Vaccine. 29 (35): 5940–9. doi:10.1016/j.vaccine.2011.06.053. PMID 21722682.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Douse CH, Green JL, Salgado PS, Simpson PJ, Thomas JC, Langsley G, Holder AA, Tate EW, Cota E (2012) Regulation of the Plasmodium motor complex: phosphorylation of Myosin A Tail Interacting Protein (MTIP) loosens its grip on MyoA. J Biol Chem
- ^ Qureshi BM, Hofmann NE, Arroyo-Olarte RD, Nickl B, Hoehne W, Jungblut PR, Lucius R, Scheerer P, Gupta N (2012) Dynein light chain 8a of Toxoplasma gondii, a unique conoid-localized β-strand-swapped homodimer, is required for an efficient parasite growth. FASEB J
- ^ Shipley K, Hekmat-Nejad M, Turner J, Moores C, Anderson R, Milligan R, Sakowicz R, Fletterick R () Structure of a kinesin microtubule depolymerization machine" EMBO J 23(7) 1422-1432
- ^ Hain AU, Weltzer RR, Hammond H, Jayabalasingham B, Dinglasan RR, Graham DR, Colquhoun DR, Coppens I, Bosch J (2012) Structural characterization and inhibition of the Plasmodium Atg8-Atg3 interaction. J Struct Biol pii: S1047-8477(12)00243-2. doi:10.1016/j.jsb.2012.09.001
- ^ Tomlins AM, Ben-Rached F, Williams RA, Proto WR, Coppens I, Ruch U, Gilberger TW, Coombs GH, Mottram JC, Müller S, Langsley (2013) Plasmodium falciparum ATG8 implicated in both autophagy and apicoplast formation" Autophagy 9(10)
- ^ Dessens JT, Saeed S, Tremp AZ, Carter V (2011) Malaria crystalloids: specialized structures for parasite transmission? Trends Parasitol 27(3) 106-110 doi:10.1016/j.pt.2010.12.004
- ^ Garnham PC, Bird RG, Baker JR (1962) Electron microscope studies of motile stages of malaria parasites. III. The ookinetes of Haemamoeba and Plasmodium" Trans R Soc Trop Med Hyg 1962;56:116–120
- ^ Carter V, Shimizu S, Arai M, Dessens JT (2008) PbSR is synthesized in macrogametocytes and involved in formation of the malaria crystalloids" Mol Microbiol 68(6) 1560-1569 doi:10.1111/j.1365-2958.2008.06254.x
- ^ Saeed S, Carter V, Tremp AZ, Dessens JT (2010) Plasmodium berghei crystalloids contain multiple LCCL proteins. Mol Biochem Parasitol 170(1) 49-53 doi:10.1016/j.molbiopara.2009.11.008
- ^ Oakley MS, Gerald N, Anantharaman V, Gao Y, Majam V, Mahajan B, Pham PT, Lotspeich-Cole L, Myers TG, McCutchan TF, Morris SL, Aravind L, Kumar S (2012) Radiation induced cellular and molecular alterations in asexual intraerythrocytic Plasmodium falciparum parasites. J Infect Dis
- ^ da Cruz LN, Juliano MA, Budu A, Juliano L, Holder AA, Blackman MJ, Garcia CR (2012) Extracellular ATP triggers proteolysis and cytosolic Ca2+ rise in Plasmodium berghei and Plasmodium yoelii malaria parasites. Malar J 11(1) 69.
- ^ Waisberg M, Cerqueira GC, Yager SB, Francischetti IM, Lu J, Gera N, Srinivasan P, Miura K, Rada B, Lukszo J, Barbian KD, Leto TL, Porcella SF, Narum DL, El-Sayed N, Miller LH, Pierce SK (2012) Plasmodium falciparum merozoite surface protein 1 blocks the proinflammatory protein S100P. Proc Natl Acad Sci USA
- ^ Gómez, ND; Safeukui, I; Adelani, AA; Tewari, R; Reddy, JK; Rao, S; Holder, A; Buffet, P; Mohandas, N; Haldar, Kasturi; et al. (2011). Spielmann, Tobias (ed.). "Deletion of a malaria invasion gene reduces death and anemia, in model hosts". PLoS ONE. 6 (9): e25477. doi:10.1371/journal.pone.0025477.
{{cite journal}}: Explicit use of et al. in:|first9=(help)CS1 maint: unflagged free DOI (link) - ^ Sakamoto H, Takeo S, Maier AG, Sattabongkot J, Cowman AF, Tsuboi T (2012). "Antibodies against a Plasmodium falciparum antigen PfMSPDBL1 inhibit merozoite invasion into human erythrocytes". Vaccine. 30 (11): 1972–80. doi:10.1016/j.vaccine.2012.01.010. PMID 22248820.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Van Tyne D, Uboldi AD, Healer J, Cowman AF, Wirth DF (2013) Modulation of PF10_0355 (MSPDBL2) alters Plasmodium falciparum response to antimalarial drugs. Antimicrob Agents Chemother
- ^ Huang YT, Lu XM, Jin XB, Zhu JY (2012) Research advances on circumsporzoite protein of Plasmodium. Zhongguo Ji Sheng Chong Xue Yu Ji Sheng Chong Bing Za Zhi 30(3) 238-242
- ^ Ding Y, Huang X, Liu T, Fu Y, Tan Z, Zheng H, Zhou T, Dai J, Xu W (2012) The Plasmodium circumsporozoite protein, a novel NF-κB inhibitor, suppresses the growth of SW480. Pathol Oncol Res
- ^ Wang J, Zhang Y, Zhao YO, Li MW, Zhang L, Dragovic S, Abraham NM, Fikrig E (2013) Anopheles gambiae circumsporozoite-protein binding-protein facilitates Plasmodium infection of mosquito salivary glands. J Infect Dis
- ^ Currà C, Pace T, Franke-Fayard BM, Picci L, Bertuccini L, Ponzi M (2011) Erythrocyte remodeling in Plasmodium berghei infection: the contribution of SEP family members. Traffic doi:10.1111/j.1600-0854.2011.01313.x
- ^ Mackellar DC, O'Neill MT, Aly AS, Sacci JB Jr, Cowman AF, Kappe SH (2010) Plasmodium falciparum PF10_0164 (ETRAMP10.3) is an essential parasitophorous vacuole and exported protein of blood stages. Eukaryotic Cell
- ^ Mackellar DC, Vaughan AM, Aly AS, Deleon S, Kappe SH (2011) A systematic analysis of the early transcribed membrane protein family throughout the life cycle of Plasmodium yoelii. Cell Microbiol. doi:10.1111/j.1462-5822.2011.01656.x
- ^ a b Currà C, Di Luca M, Picci L, de Sousa Silva Gomes Dos Santos C, Siden-Kiamos I, Pace T, Ponzi M (2013) The ETRAMP family member SEP2 is expressed throughout Plasmodium berghei life cycle and is released during sporozoite gliding motility" PLoS One 8(6) e67238. doi:10.1371/journal.pone.0067238
- ^ Taechalertpaisarn T, Crosnier C, Bartholdson SJ, Hodder AN, Thompson J, Bustamante LY, Wilson DW, Sanders PR, Wright GJ, Rayner JC, Cowman AF, Gilson PR, Crabb BS (2012) Biochemical and functional analysis of two Plasmodium falciparum blood-stage 6-cys proteins: P12 and P41" PLoS One 7(7) e41937.
- ^ Elliott JF, Albrecht GR, Gilladoga A, Handunnetti SM, Neequaye J, Lallinger G, Minjas JN, Howard RJ (1990) Genes for Plasmodium falciparum surface antigens cloned by expression in COS cells" Proc Natl Acad Sci USA 87(16) 6363-6367
- ^ Saeed S, Tremp AZ, Dessens JT (2012) Conformational co-dependence between Plasmodium berghei LCCL proteins promotes complex formation and stability. Mol Biochem Parasitol
- ^ Saeed S, Carter V, Tremp AZ, Dessens JT (2013) Translational repression controls temporal expression of the Plasmodium berghei LCCL protein complex. Mol Biochem Parasitol pii: S0166-6851(13)00053-4. doi:10.1016/j.molbiopara.2013.04.006
- ^ Mattei D, Scherf A (1992) The Pf332 gene codes for a megadalton protein of Plasmodium falciparum asexual blood stages. Mem Inst Oswaldo Cruz 87: 163–168
- ^ Moll K, Chene A, Ribacke U, Kaneko O, Nilsson S et al (2007) A novel DBL-domain of the P. falciparum 332 molecule possibly involved in erythrocyte adhesion" PLoS ONE 2: e477
- ^ a b Hinterberg K, Scherf A, Gysin J, Toyoshima T, Aikawa M et al (1994) Plasmodium falciparum: the Pf332 antigen is secreted from the parasite by a brefeldin A dependent pathway and is translocated to the erythrocyte membrane via the Maurer's clefts. Exp Parasitol 79: 279–291
- ^ Pavithra SR, Kumar R, Tatu U (2007) Systems analysis of chaperone networks in the malarial parasite Plasmodium falciparum" PLoS Comput Biol 3: 1701–1715
- ^ a b Nilsson S, Angeletti D, Wahlgren M, Chen Q, Moll K (2012) Plasmodium falciparum antigen 332 is a resident peripheral membrane protein of Maurer's clefts" PLoS One 7(11) e46980. doi:10.1371/journal.pone.0046980
- ^ Hodder AN, Maier AG, Rug M, Brown M, Hommel M et al (2009) Analysis of structure and function of the giant protein Pf332 in Plasmodium falciparum" Mol Microbiol 71: 48–65
- ^ Waller KL, Stubberfield LM, Dubljevic V, Buckingham DW, Mohandas N et al (2010) Interaction of the exported malaria protein Pf332 with the red blood cell membrane skeleton" Biochim Biophys Acta 1798: 861–871 doi:10.1016/j.bbamem.2010.01.018
- ^ Prajapati SK, Singh OP (2013) Remodeling of human red cells infected with Plasmodium falciparum and the impact of PHIST proteins. Blood Cells Mol Dis pii: S1079-9796(13)00149-6. doi:10.1016/j.bcmd.2013.06.003
- ^ a b c Uchime O, Herrera R, Reiter K, Kotova S, Shimp RL Jr, Miura K, Jones D, Lebowitz J, Ambroggio X, Hurt DE, Jin AJ, Long C, Miller LH, Narum DL (2012) Analysis of the conformation and function of the Plasmodium falciparum merozoite proteins MTRAP and PTRAMP. Eukaryot Cell
- ^ Tham, WH; Wilson, DW; Lopaticki, S; Schmidt, CQ; Tetteh-Quarcoo, PB; Barlow, PN; Richard, D; Corbin, JE; Beeson, JG; et al. (2010). "Complement receptor 1 is the host erythrocyte receptor for Plasmodium falciparum PfRh4 invasion ligand". Proc Natl Acad Sci U S A. 107 (40): 17327–17332. doi:10.1073/pnas.1008151107. PMC 2951459. PMID 20855594.
{{cite journal}}: Explicit use of et al. in:|first9=(help) - ^ Kilili GK, Lacount DJ (2011). "An erythrocyte cytoskeleton-binding motif in exported Plasmodium falciparum proteins". Eukaryotic Cell. 10 (11): 1439. doi:10.1128/EC.05180-11.
- ^ Diez-Silva M, Park Y, Huang S, Bow H, Mercereau-Puijalon O, Deplaine G, Lavazec C, Perrot S, Bonnefoy S, Feld MS, Han J, Dao M, Suresh S (2012) Pf155/RESA protein influences the dynamic microcirculatory behavior of ring-stage Plasmodium falciparum infected red blood cells. Sci Rep 2:614
- ^ Gilson PR, Nebl T, Vukcevic D, Moritz RL, Sargeant T, Speed TP, Schofield L, Crabb BS (2006) Identification and stoichiometry of glycosylphosphatidylinositolanchored membrane proteins of the human malaria parasite Plasmodium falciparum. Mol Cell Proteomics 5:1286-1299
- ^ Sakura T, Yahata K, Kaneko O (2012) The upstream sequence segment of the C-terminal cysteine-rich domain is required for microneme trafficking of Plasmodium falciparum erythrocyte binding antigen 175. Parasitol Int. 2012 Dec 22. pii: S1383-5769(12)00162-6. doi:10.1016/j.parint.2012.12.002
- ^ Ghosh AK, Jacobs-Lorena M (2011) Surface-expressed enolases of Plasmodium and other pathogens. Mem Inst Oswaldo Cruz 106(Suppl 1) 85-90
- ^ Schlarman, MS; Roberts, RN; Kariuki, MM; Lacrue, AN; Ou, R; Beerntsen, BT (2012). "PFE0565w, a Plasmodium falciparum protein Expressed in salivary gland sporozoites". Am J Trop Med Hyg. 86 (6): 943–954. doi:10.4269/ajtmh.2012.11-0797. PMC 3366537. PMID 22665598.
- ^ Xangsayarath P, Kaewthamasorn M, Yahata K, Nakazawa S, Sattabongkot J, Udomsangpetch R, Kaneko O (2012) Positive diversifying selection on the Plasmodium falciparum surf gene in Thailand. Trop Med Health 40(3) 79-89 doi:10.2149/tmh.2012-12
- ^ Valle-Delgado JJ, Urbán P, Fernàndez-Busquets X (2013) Demonstration of specific binding of heparin to Plasmodium falciparum-infected vs. non-infected red blood cells by single-molecule force spectroscopy. Nanoscale
- ^ Lalle M, Curra C, Ciccarone F, Pace T, Cecchetti S, Fantozzi L, Ay B, Braun Breton C, Ponzi M. (2010). "Dematin, a component of the erythrocyte membrane-skeleton, is internalized by the malaria parasite and associates with Plasmodium 14-3-3". J. Biol. Chem.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Sicard A, Semblat JP, Doerig C, Hamelin R, Moniatte M, Dorin-Semblat D, Spicer JA, Srivastava A, Retzlaff S, Heussler V, Waters AP, Doerig C (2011). "Activation of a PAK-MEK signalling pathway in malaria parasite-infected erythrocytes". Cell Microbiol. 13 (6): 836–45. doi:10.1111/j.1462-5822.2011.01582.x. PMC 3123749. PMID 21371233.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Bagnaresi P, de Barros NM, Assis DM, Melo PM, Fonseca RG, Juliano MA, Pesquero JB, Juliano L, Rosenthal PJ, Carmona AK, Gazarini ML (2012) Intracellular proteolysis of kininogen by malaria parasites promotes release of active kinins. Malar J 11(1) 156
- ^ Koncarevic S, Rohrbach P, Deponte M, Krohne G, Prieto JH, Yates J 3rd, Rahlfs S, Becker K (2009) The malarial parasite Plasmodium falciparum imports the human protein peroxiredoxin 2 for peroxide detoxification" Proc Natl Acad Sci USA 106(32) 13323-8. doi:10.1073/pnas.0905387106
- ^ Bonday ZQ, Taketani S, Gupta PD, Padmanaban G (1997) Heme biosynthesis by the malarial parasite. Import of delta-aminolevulinate dehydrase from the host red cell" J Biol Chem 272(35) 21839-21846
- ^ a b Spillman NJ, Allen RJ, Kirk K (2013) Na+ extrusion imposes an 'acid load' on the intraerythrocytic malaria parasite. Mol Biochem Parasitol. 2013 Apr 23. pii: S0166-6851(13)00051-0. doi:10.1016/j.molbiopara.2013.04.004 Cite error: The named reference "Spillman2013" was defined multiple times with different content (see the help page).
- ^ Henry RI, Cobbold SA, Allen RJ, Khan A, Hayward R, Lehane AM, Bray PG, Howitt SM, Biagini GA, Saliba KJ, Kirk K (2010). "An acid-loading chloride transport pathway in the intraerythrocytic malaria parasite, Plasmodium falciparum". J. Biol. Chem. 285 (24): 18615–26. doi:10.1074/jbc.M110.120980. PMC 2881787. PMID 20332090.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Müller IB, Knöckel J, Eschbach ML, Bergmann B, Walter RD, Wrenger C (2010) Secretion of an acid phosphatase provides a possible mechanism to acquire) host nutrients by Plasmodium falciparum. Cell Microbiol.
- ^ Cobbold SA, Martin RE, Kirk K (2010). "Methionine transport in the malaria parasite Plasmodium falciparum". Parasitol.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Sinou V, Quang LH, Pelleau S, Huong VN, Huong NT, Tai LM, Bertaux L, Desbordes M, Latour C, Long LQ, Thanh NX, Parzy D (2011) Polymorphism of Plasmodium falciparum Na+/H+ exchanger is indicative of a low in vitro quinine susceptibility in isolates from Viet Nam. Malar J. 2011 Jun 14;10(1) 164
- ^ Kone A, Mu J, Maiga H, Beavogui AA, Yattara O, Sagara I, Tekete MM, Traore OB, Dara A, Dama S, Diallo N, Kodio A, Traoré A, Björkman A, Gil JP, Doumbo OK, Wellems TE, Djimde AA (2012) Quinine treatment selects Pfnhe1 ms47601 polymorphism in Malian patients with falciparum malaria. J Infect Dis
- ^ Salcedo-Sora JE, Ochong E, Beveridge S, Johnson D, Nzila A, Biagini GA, Stocks PA, O'Neill PM, Krishna S, Bray PG, Ward SA (2011). "The molecular basis of folate salvage in Plasmodium falciparum: Characterization of two folate transporters". J Biol Chem. 286 (52): 44659–68. doi:10.1074/jbc.M111.286054. PMC 3247980. PMID 21998306.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Wang P, Wang Q, Sims PF, Hyde JE (2009) Characterisation of exogenous folate transport in Plasmodium falciparum. Mol Biochem Parasitol 154(1) 40-51
- ^ Riegelhaupt PM, Frame IJ, Akabas MH (2010) Transmembrane segment 11 appears to line the purine permeation pathway of the Plasmodium falciparum equilibrative nucleoside transporter 1 (PfENT1). J Biol Chem.
- ^ El Bissati K, Zufferey R, Witola WH, Carter NS, Ullman B, Ben Mamoun C (2006) The plasma membrane permease PfNT1 is essential for purine salvage in the human malaria parasite Plasmodium falciparum" Proc Natl Acad Sci USA 103(24) 9286-9291
- ^ Quashie NB, Ranford-Cartwright LC, De Koning HP (2010) Uptake of purines in Plasmodium falciparum-infected human erythrocytes is mostly mediated by the human equilibrative nucleoside transporter and the human Facilitative Nucleobase Transporter. Malar J 9(1) 36
- ^ Downie MJ, El Bissati K, Bobenchik AM, Nic Lochlainn L, Amerik A, Zufferey R, Kirk K, Ben Mamoun C (2010). "PfNT2: a permease of the equilibrative nucleoside transporter family in the endoplasmic reticulum of Plasmodium falciparum". J Biol Chem. 285 (27): 20827–33. doi:10.1074/jbc.M110.118489. PMC 2898299. PMID 20439460.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Frame IJ, Merino EF, Schramm VL, Cassera MB, Akabas MH (2012) Malaria parasite type 4 equilibrative nucleoside transporters (ENT4) are purine transporters with distinct substrate specificity. Biochem J
- ^ Niemand J, Louw AI, Birkholtz L, Kirk K (2012) Polyamine uptake by the intraerythrocytic malaria parasite, Plasmodium falciparum. Int J Parasitol
- ^ Nguitragool W, Bokhari AA, Pillai AD, Rayavara K, Sharma P, Turpin B, Aravind L, Desai S (2011). "Malaria parasite clag genes determine nutrient uptake channel activity on infected red blood cells". Cell. 145 (5): 665–677. doi:10.1016/j.cell.2011.05.002. PMC 3105333. PMID 21620134.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Pillai AD, Nguitragool W, Lyko B, Dolinta K, Butler MM, Nguyen ST, Peet NP, Bowlin TL, Desai SA (2012) Solute restriction reveals an essential role for clag3-associated channels in malaria parasite nutrient acquisition. Mol Pharmacol
- ^ Alkhalil A, Hong L, Nguitragool W, Desai SA (2011). "Voltage-dependent inactivation of the plasmodial surface anion channel via a cleavable cytoplasmic component". Biochim Biophys Acta.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Alexandre JS, Xangsayarath P, Kaewthamasorn M, Yahata K, Sattabongkot J, Udomsangpetch R, Kaneko O (2012) Stable allele frequency distribution of the Plasmodium falciparum clag genes encoding components of the high molecular weight rhoptry protein complex. Trop Med Health 40(3) 71-77 doi:10.2149/tmh.2012-13
- ^ a b Sharma P, Wollenberg K, Sellers M, Zainabadi K, Galinsky K, Moss E, Nguitragool W, Neafsey D, Desai SA (2013) An epigenetic antimalarial resistance mechanism involving parasite genes linked to nutrient uptake. J Biol Chem
- ^ Mira-Martínez S, Rovira-Graells N, Crowley VM, Altenhofen LM, Llinás M, Cortés A (2013) Epigenetic switches in clag3 genes mediate blasticidin S resistance in malaria parasites. Cell Microbiol doi:10.1111/cmi.12162
- ^ Alexandre JS, Kaewthamasorn M, Yahata K, Nakazawa S, Kaneko O (2011) Positive selection on the Plasmodium falciparum clag2 gene encoding a component of the erythrocyte-binding rhoptry protein complex. Trop Med Health 39(3) 77-82
- ^ Chen LY (2012) Glycerol inhibits water permeation through Plasmodium falciparum aquaglyceroporin. J Struct Biol pii: S1047-8477(12)00289-4. doi:10.1016/j.jsb.2012.10.007
- ^ Choveaux DL, Przyborski JM, Goldring JD (2012) A Plasmodium falciparum copper-binding membrane protein with copper transport motifs. Malar J 11(1) 397
- ^ Augagneur Y, Jaubert L, Schiovani M, Pachikara N, Garg A, Usmani-Brown S, Wesolowski D, Zeller S, Ghosal A, Cornillot E, Said HM, Kumar P, Altman S, Ben Mamoun C (2013) Identification and functional analysis of the primary pantothenate transporter, PfPAT, of the human malaria parasite Plasmodium falciparum. J Biol Chem
- ^ Riglar DT, Rogers KL, Hanssen E, Turnbull L, Bullen HE, Charnaud SC, Przyborski J, Gilson PR, Whitchurch CB, Crabb BS, Baum J, Cowman AF (2013) Spatial association with PTEX complexes defines regions for effector export into Plasmodium falciparum-infected erythrocytes. Nat Commun 4:1415. doi:10.1038/ncomms2449
- ^ de Koning-Ward TF, Gilson PR, Boddey JA, Rug M, Smith BJ, Papenfuss AT, Sanders PR, Lundie RJ, Maier AG, Cowman AF, Crabb BS (2009) A newly discovered protein export machine in malaria parasites" Nature 459(7249) 945-9. doi:10.1038/nature08104
- ^ a b c Matz JM, Matuschewski K, Kooij TW (2013) Two putative protein export regulators promote malaria blood stage development in vivo. Mol Biochem Parasitol pii: S0166-6851(13)00134-5. doi:10.1016/j.molbiopara.2013.09.003
- ^ Sharma A, Sharma A, Dixit S, Sharma A (2011) Structural insights into thioredoxin-2: a component of malaria parasite protein secretion machinery. Sci Rep 1:179
- ^ Rached FB, Ndjembo-Ezougou C, Chandran S, Talabani H, Yera H, Dandavate V, Bourdoncle P, Meissner M, Tatu U, Langsley G (2011) Construction of a Plasmodium falciparum Rab-interactome identifies CK1 and PKA as Rab-effector kinases in malaria parasites. Biol Cell doi:10.1111/boc.201100081
- ^ Tarr SJ, Cryar A, Thalassinos K, Haldar K, Osborne AR (2012) The C-terminal portion of the cleaved HT motif is necessary and sufficient to mediate export of proteins from the malaria parasite into its host cell. Mol Microbiol doi:10.1111/mmi.12133
- ^ Henriques G, Martinelli A, Rodrigues L, Modrzynska K, Fawcett R, Houston DR, Borges ST, D Alessandro U, Tinto H, Karema C, Hunt P, Cravo P (2013) Artemisinin resistance in rodent malaria - mutation in the AP2 adaptor mu-chain suggests involvement of endocytosis and membrane protein trafficking. Malar J 12(1) 118
- ^ Haase S, Hanssen E, Matthews K, Kalanon M, de Koning-Ward TF (2013) The exported protein PbCP1 localises to cleft-like structures in the rodent malaria parasite Plasmodium berghei" PLoS One 8(4) e61482. doi:10.1371/journal.pone.0061482
- ^ Parish LA, Mai DW, Jones ML, Kitson EL, Rayner JC (2013) A member of the Plasmodium falciparum PHIST family binds to the erythrocyte cytoskeleton component band 4.1.Malar J 12(1) 160
- ^ Weng H, Guo X, Papoin J, Wang J, Coppel R, Mohandas N, An X (2013) Interaction of Plasmodium falciparum Knob-associated Histidine-rich Protein (KAHRP) with erythrocyte ankyrin R is required for its attachment to the erythrocyte membrane. Biochim Biophys Acta pii: S0005-2736(13)00329-5. doi:10.1016/j.bbamem.2013.09.014
- ^ Talevich E, Tobin AB, Kannan N, Doerig C (2012) An evolutionary perspective on the kinome of malaria parasites" Philos Trans R Soc Lond B Biol Sci 367(1602) 2607-2618
- ^ Talevich E, Mirza A, Kannan N (2011). "Structural and evolutionary divergence of eukaryotic protein kinases in Apicomplexa". BMC Evol Biol. 11 (1): 321. doi:10.1186/1471-2148-11-321.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Trivedi V, Nag S (2012) In silico characterization of atypical kinase, PFD0975w from Plasmodium kinome: A suitable target for drug discovery. Chem Biol Drug Des doi:10.1111/j.1747-0285.2012.01321.x.
- ^ Chouhan DK, Sharon A, Bal C (2012) Molecular and structural insight into plasmodium falciparum RIO2 kinase. J Mol Model
- ^ Nag S, Prasad KM, Bhowmick A, Deshmukh R, Trivedi V (2012) PfRIO-2 kinase is a potential therapeutic target of antimalarial protein kinase inhibitors. Curr Drug Discov Technol
- ^ Agarwal S, Kern S, Halbert J, Przyborski JM, Baumeister S, Dandekar T, Doerig C, Pradel G. (2011) Two nucleus-localized CDK-like kinases with crucial roles for malaria parasite erythrocytic replication are involved in phosphorylation of splicing factor. J. Cell. Biochem. doi:10.1002/jcb.23034
- ^ Dorin-Semblat D; Schmitt S; Semblat JP; Sicard A; Reininger L; Goldring D; Patterson S; Quashie N; Chakrabarti D (2011). "Plasmodium falciparum NIMA-related kinase Pfnek-1: sex specificity and assessment of essentiality for the erythrocytic asexual cycle". Microbiology. 157 (10): 2785–2794. doi:10.1099/mic.0.049023-0.
{{cite journal}}:|first10=missing|last10=(help);|first11=missing|last11=(help); Invalid|display-authors=9(help); Unknown parameter|author-separator=ignored (help)CS1 maint: unflagged free DOI (link) - ^ Low H, Chua CS, Sim TS (2011). "Plasmodium falciparum possesses a unique dual-specificity serine/threonine and tyrosine kinase, Pfnek3". Cell Mol Life Sci.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Reininger L, Garcia M, Tomlins A, Müller S, Doerig C (2012) The Plasmodium falciparum, Nima-related kinase Pfnek-4: a marker for asexual parasites committed to sexual differentiation. Malar J. 11(1) 250.
- ^ Ma J, Rahlfs S, Jortzik E, Heiner Schirmer R, Przyborski J, Becker K (2012) Subcellular localization of adenylate kinases in Plasmodium falciparum. FEBS Lett
- ^ Law AW, Lescar J, Hao Q, Kotaka M (2012) Expression, purification, crystallization and preliminary X-ray analysis of Plasmodium falciparum GTP:AMP phosphotransferase. Acta Crystallogr Sect F Struct Biol Cryst Commun 68(6) 671-674
- ^ Lauciello L, Kappes B, Scapozza L, Perozzo R (2013) Expression, purification and biochemical characterization of recombinant Ca-dependent protein kinase 2 of the malaria parasite Plasmodium falciparum. Protein Expr Purif pii: S1046-5928(13)00112-5. doi:10.1016/j.pep.2013.06.006
- ^ Azevedo MF, Sanders PR, Krejany E, Nie CQ, Fu P, Bach LA, Wunderlich G, Crabb BS, Gilson PR (2013) Inhibition of Plasmodium falciparum CDPK1 by conditional expression of its J-domain demonstrates a key role in schizont development. Biochem J
- ^ Bansal A, Singh S, More KR, Hans D, Nangalia K, Yogavel M, Sharma A, Chitnis CE (2013) Characterization of Plasmodium falciparum calcium-dependent protein kinase 1 (PfCDPK1) and its role in microneme secretion during erythrocyte invasion. J Biol Chem. 2013 Jul 5;288(27) 19643. doi:10.1074/jbc.A112.411934
- ^ Holder AA, Mohd Ridzuan MA, Green JL (2012) Calcium dependent protein kinase 1 and calcium fluxes in the malaria parasite" Microbes Infect 14(10) 825-30. doi:10.1016/j.micinf.2012.04.006
- ^ Ahmed A, Gaadhe K, Sharma GP, Kumar N, Neculai M, Hui R, Mohanty D, Sharma P (2012) Novel insights into the regulation of malarial calcium-dependent protein kinase 1" FASEB J 26(8) 3212-21. doi:10.1096/fj.12-203877
- ^ Möskes C, Burghaus PA, Wernli B, Sauder U, Dürrenberger M, Kappes B (2004) Export of Plasmodium falciparum calcium-dependent protein kinase 1 to the parasitophorous vacuole is dependent on three N-terminal membrane anchor motifs" Mol Microbiol 54(3) 676-691
- ^ McCoy JM, Whitehead L, van Dooren GG, Tonkin CJ (2012) TgCDPK3 regulates calcium-dependent egress of Toxoplasma gondii from host cells" PLoS Pathog 8(12) e1003066. doi:10.1371/journal.ppat.1003066
- ^ a b Kato N, Sakata T, Breton G, Le Roch KG, Nagle A, et al (2008) Gene expression signatures and small-molecule compounds link a protein kinase to Plasmodium falciparum motility. Nat Chem Biol 4: 347–356
- ^ Green JL, Rees-Channer RR, Howell SA, Martin SR, Knuepfer E, et al (2008) The motor complex of Plasmodium falciparum: phosphorylation by a calcium-dependent protein kinase" J Biol Chem 283: 30980–30989.
- ^ Sebastian S, Brochet M, Collins MO, Schwach F, Jones ML, Goulding D, Rayner JC, Choudhary JS, Billker O (2012) A Plasmodium calcium-dependent protein kinase controls zygote development and transmission by translationally activating repressed mRNAs. Cell Host Microbe 12(1) 9-19
- ^ Dvorin JD, Martyn DC, Patel SD, Grimley JS, Collins CR, (2010) A plant-like kinase in Plasmodium falciparum regulates parasite egress from erythrocytes" Science 328: 910–912
- ^ Ishino T, Orito Y, Chinzei Y, Yuda M (2006) A calcium-dependent protein kinase regulates Plasmodium ookinete access to the midgut epithelial cell" Mol Microbiol 59: 1175–1184
- ^ Billker O, Dechamps S, Tewari R, Wenig G, Franke-Fayard B, et al. (2004) Calcium and a calcium-dependent protein kinase regulate gamete formation and mosquito transmission in a malaria parasite" Cell 117: 503–514
- ^ Ojo KK, Eastman RT, Vidadala R, Zhang Z, Rivas KL, Choi R, Lutz JD, Reid MC, Fox AM, Hulverson MA, Kennedy M, Isoherranen N, Kim LM, Comess KM, Kempf DJ, Verlinde CL, Su XZ, Kappe S, Maly DJ, Fan E, Van Voorhis WC (2013) A specific inhibitor of PfCDPK4 blocks malaria transmission: Chemical-genetic validation. J Infect Dis
- ^ Lin DT, Goldman ND, Syin C (1996) Stage-specific expression of a Plasmodium falciparum protein related to the eukaryotic mitogen-activated protein kinases. Mol Biochem Parasitol 78(1-2) 67-77
- ^ Wierk JK, Langbehn A, Kamper M, Richter S, Burda PC, Heussler VT, Deschermeier C (2013) Plasmodium berghei MAPK1 displays differential and dynamic subcellular localizations during liver stage development" PLoS One 8(3) e59755. doi:10.1371/journal.pone.0059755
- ^ Dorin D, Alano P, Boccaccio I, Cicéron L, Doerig C, Sulpice R, Parzy D, Doerig C (1999) An atypical mitogen-activated protein kinase (MAPK) homologue expressed in gametocytes of the human malaria parasite Plasmodium falciparum. Identification of a MAPK signature" J Biol Chem 74(42) 29912-29920
- ^ Brandt GS, Bailey S (2013) Dematin, a human erythrocyte cytoskeletal protein, is a substrate for a recombinant FIKK kinase from Plasmodium falciparum. Mol Biochem Parasitol. 2013 Aug 21. pii: S0166-6851(13)00130-8 doi:10.1016/j.molbiopara.2013.08.003
- ^ a b Dastidar EG, Dayer G, Holland ZM, Dorin-Semblat D, Claes A, Chene A, Sharma A, Hamelin R, Moniatte M, Lopez-Rubio JJ, Scherf A, Doerig C (2012) Involvement of Plasmodium falciparum protein kinase CK2 in the chromatin assembly pathway. BMC Biol 10(1) 5 Cite error: The named reference "Dastidar2012" was defined multiple times with different content (see the help page).
- ^ Hopp CS, Flueck C, Solyakov L, Tobin A, Baker DA (2012) Spatiotemporal and functional characterisation of the Plasmodium falciparum cGMP-dependent protein kinase" PLoS One 7(11) e48206. doi:10.1371/journal.pone.0048206
- ^ Spry C, Macuamule C, Lin Z, Virga KG, Lee RE, Strauss E, Saliba KJ (2013) Pantothenamides are potent, on-target inhibitors of Plasmodium falciparum growth when serum pantetheinase is inactivated" PLoS One 8(2) e54974 doi:10.1371/journal.pone.0054974
- ^ Sherrer RL, O'Donoghue P, Söll D (2008) Characterization and evolutionary history of an archaeal kinase involved in selenocysteinyl-tRNA formation" Nucleic Acids Res 36(4) 1247-59. doi:10.1093/nar/gkm1134
- ^ Sharma A, Biswas S (2005) Stage-specific cytosolic protein kinase C-like activity in human malarial parasite Plasmodium falciparum. Indian J Biochem Biophys. 2005 Jun;42(3) 145-151
- ^ Abdi AI, Carvalho TG, Wilkes JM, Doerig C (2013) A secreted Plasmodium falciparum kinase reveals a signature motif for classification of tyrosine kinase-like kinases. Microbiology
- ^ Dorin-Semblat D, Sicard A, Doerig C, Ranford-Cartwright L, Doerig C (2008) Disruption of the PfPK7 gene impairs schizogony and sporogony in the human malaria parasite Plasmodium falciparum. Eukaryot Cell 7(2) 279-285
- ^ Wilkes JM, Doerig C (2008) The protein-phosphatome of the human malaria parasite Plasmodium falciparum" BMC Genomics 9: 412
- ^ Patzewitz EM, Guttery DS, Poulin B, Ramakrishnan C, Ferguson DJ, Wall RJ, Brady D, Holder AA, Szöőr B, Tewari R (2013) An ancient protein phosphatase, SHLP1, is critical to microneme development in Plasmodium ookinetes and parasite transmission. Cell Rep pii: S2211-1247(13)00056-9. doi:10.1016/j.celrep.2013.01.032
- ^ Balu B, Campbell C, Sedillo J, Maher S, Singh N, Thomas P, Zhang M, Pance A, Otto TD, Rayner JC, Adams JH (2013) An atypical MAPK phosphatase implicated in regulating transition from pre-S-phase asexual intraerythrocytic development of Plasmodium falciparum. Eukaryot Cell
- ^ Freville A, Landrieu I, Garcia-Gimeno MA, Vicogne J, Montbarbon M, Bertin B, Verger A, Kalamou H, Sanz P, Werkmeister E, Pierrot C, Khalife J (2011). "Plasmodium falciparum inhibitor 3 homolog increases protein phosphatase type 1 activity and is essential for parasitic survival". J Biol Chem.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Frèville A, Cailliau-Maggio K, Pierrot C, Tellier G, Kalamou H, Lafitte S, Martoriati A, Pierce RJ, Bodart JF, Khalife J (2013) Plasmodium falciparum encodes a conserved active inhibitor-2 for protein phosphatase type 1: perspectives for novel anti-plasmodial therapy. BMC Biol 11(1) 80
- ^ Rosenthal PJ (2011). "Falcipains and other cysteine proteases of malaria parasites". Adv Exp Med Biol. Advances in Experimental Medicine and Biology. 712: 30–48. doi:10.1007/978-1-4419-8414-2_3. ISBN 978-1-4419-8413-5. PMID 21660657.
- ^ Armada A, Gazarini M, Gonçalves LM, Antunes S, Custódio A, Rodrigues A, Almeida AJ, Silveira H, Rosário VD, Santos-Gomes G, Domingos A (2013) Generation of an antibody that recognizes Plasmodium chabaudi cysteine protease (chabaupain-1) in both sexual and asexual parasite life cycle and evaluation of chabaupain-1 vaccine potential. Exp Parasitol pii: S0014-4894(13)00167-7. doi:10.1016/j.exppara.2013.06.009
- ^ a b Tanaka TQ, Deu E, Molina-Cruz A, Ashburne MF, Ali O, Suri A, Kortagere S, Bogyo M, Williamson KC (2013) Plasmodium dipeptidyl aminopeptidases as malaria transmission blocking drug targets. Antimicrob Agents Chemother Cite error: The named reference "Tanaka2013" was defined multiple times with different content (see the help page).
- ^ Arisue N, Hirai M, Arai M, Matsuoka H, Horii T (2007) Phylogeny and evolution of the SERA multigene family in the genus Plasmodium" J Mol Evol 65:82–91
- ^ Soh BY, Song HO, Lee Y, Lee J, Kaewintajuk K, Lee B, Choi YY, Cho JH, Choi S, Park H (2013) Identification of active Plasmodium falciparum calpain to establish screening system for Pf-calpain-based drug development. Malar J 12(1) 47
- ^ Mitchell D, Bell A (2003) PEST sequences in the malaria parasite Plasmodium falciparum: a genomic study. Malar J 2:16
- ^ Pei Y, Miller JL, Lindner SE, Vaughan AM, Torii M, Kappe SH (2013) Plasmodium yoelii inhibitor of cysteine proteases is exported to exomembrane structures and interacts with yoelipain-2 during asexual blood stage development. Cell Microbiol doi:10.1111/cmi.12124
- ^ Prasad R, Atul, Soni A, Puri SK, Sijwali PS (2012) Expression, characterization, and cellular localization of knowpains, papain-like cysteine proteases of the Plasmodium knowlesi malaria parasite" PLoS One 7(12) e51619. doi:10.1371/journal.pone.0051619
- ^ Ragheb D, Dalal S, Bompiani KM, Ray WK, Klemba M (2011). "Distribution and biochemical properties of an M1-family aminopeptidase in Plasmodium falciparum indicate a role in vacuolar hemoglobin catabolism". J Biol Chem. 286 (31): 27255–65. doi:10.1074/jbc.M111.225318. PMC 3149319. PMID 21659511.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Harbut MB, Velmourougane G, Dalal S, Reiss G, Whisstock JC, Onder O, Brisson D, McGowan S, Klemba M, Greenbaum DC (2011). "Bestatin-based chemical biology strategy reveals distinct roles for malaria M1- and M17-family aminopeptidases". Proc Natl Acad Sci USA. 108 (34): E526–34. doi:10.1073/pnas.1105601108. PMC 3161592. PMID 21844374.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Sivaraman KK, Oellig CA, Huynh K, Atkinson SC, Poreba M, Perugini MA, Trenholme KR, Gardiner DL, Salvesen G, Drag M, Dalton JP, Whisstock JC, McGowan S (2012) X-ray crystal structure and specificity of the Plasmodium falciparum malaria aminopeptidase PfM18AAP. J Mol Biol
- ^ Lhouvum K, Ramakrishnan V, Trivedi V (2013) Insight into structural and biochemical determinants of substrate specificity of PFI1625c: Correlation analysis of protein-peptide molecular models. J Mol Graph Model 43C:21-30. doi:10.1016/j.jmgm.2013.03.008
- ^ Gupta D, Yedidi RS, Varghese S, Kovari LC, Woster PM (2010). "Mechanism-based inhibitors of the aspartyl protease plasmepsin II as potential antimalarial agents". J. Med. Chem. 53 (10): 4234–47. doi:10.1021/jm100233b. PMID 20438064.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Bhaumik P, Xiao H, Hidaka K, Gustchina A, Kiso Y, Yada RY, Wlodawer A (2011) Structural insights into activation and inhibition of histo-aspartic protease (HAP) from Plasmodium falciparum. Biochemistry
- ^ Bhaumik P, Gustchina A, Wlodawer A (2012) Structural studies of vacuolar plasmepsins" Biochim Biophys Acta 1824(1) 207-23. doi:10.1016/j.bbapap.2011.04.008
- ^ Moura PA, Dame JB, Fidock DA (2009) Role of Plasmodium falciparum digestive vacuole plasmepsins in the specificity and antimalarial mode of action of cysteine and aspartic protease inhibitors" Antimicrob Agents Chemother 53(12) 4968-4978
- ^ Banerjee R, Francis SE, Goldberg DE (2003) Food vacuole plasmepsins are processed at a conserved site by an acidic convertase activity in Plasmodium falciparum. Mol Biochem Parasitol 129(2) 157-165
- ^ Kim YM, Lee MH, Piao TG, Lee JW, Kim JH, Lee S, Choi KM, Jiang JH, Kim TU, Park H (2006) Prodomain processing of recombinant plasmepsin II and IV, the aspartic proteases of Plasmodium falciparum, is auto- and trans-catalytic. J Biochem 139(2) 189-195
- ^ Klemba M, Goldberg DE (2005) Characterization of plasmepsin V, a membrane-bound aspartic protease homolog in the endoplasmic reticulum of Plasmodium falciparum. Mol Biochem Parasitol 143(2) 183-191
- ^ Boddey JA, Carvalho TG, Hodder AN, Sargeant TJ, Sleebs BE, Marapana D, Lopaticki S, Nebl T, Cowman AF (2013) Role of Plasmepsin V in export of diverse protein families from the Plasmodium falciparum exportome. Traffic doi:10.1111/tra.12053
- ^ Marapana DS, Wilson DW, Zuccala ES, Dekiwadia CD, Beeson JG, Ralph SA, Baum J (2012) Malaria parasite signal peptide peptidase is an ER-resident protease required for growth but not invasion. Traffic doi:10.1111/j.1600-0854.2012.01402.x
- ^ Ponder EL; Albrow VE; Leader BA; Békés M; Mikolajczyk J; Fonović UP; Shen A; Drag M; Xiao J (2011). "Functional characterization of a SUMO deconjugating protease of Plasmodium falciparum using newly identified small molecule inhibitors". Chem Biol. 18 (6): 711–721. doi:10.1016/j.chembiol.2011.04.010. PMC 3131532. PMID 21700207.
{{cite journal}}:|first10=missing|last10=(help);|first11=missing|last11=(help); Invalid|display-authors=9(help); Unknown parameter|author-separator=ignored (help); no-break space character in|first11=at position 4 (help) - ^ Alam A, Bhatnagar RK, Chauhan VS (2012) Expression and characterization of catalytic domain of Plasmodium falciparum subtilisin-like protease 3. Mol Biochem Parasitol
- ^ Tawk L, Lacroix C, Gueirard P, Kent R, Gorgette O, Thiberge S, Mercereau-Puijalon O, Ménard R, Barale JC (2013) A key role for Plasmodium subtilisin-like SUB1 in egress of malaria parasites from host hepatocytes. J Biol Chem
- ^ Yeoh S, O'Donnell RA, Koussis K, Dluzewski AR, Ansell KH, Osborne SA, Hackett F, Withers-Martinez C, Mitchell GH, Bannister LH, Bryans JS, Kettleborough CA, Blackman MJ (2007) Subcellular discharge of a serine protease mediates release of invasive malaria parasites from host erythrocytes" Cell 131(6) 1072-1083
- ^ Agarwal S, Singh MK, Garg S, Chitnis CE, Singh S (2013) Ca(2+)-mediated exocytosis of subtilisin-like protease 1: a key step in egress of Plasmodium falciparum merozoites. Cell Microbiol 15(6) 910-921. doi:10.1111/cmi.12086
- ^ Koussis K, Withers-Martinez C, Yeoh S, Child M, Hackett F, Knuepfer E, Juliano L, Woehlbier U, Bujard H, Blackman MJ (2009) A multifunctional serine protease primes the malaria parasite for red blood cell invasion" EMBO J 28(6) 725-735. doi:10.1038/emboj.2009.22
- ^ Harris PK, Yeoh S, Dluzewski AR, O'Donnell RA, Withers-Martinez C, Hackett F, Bannister LH, Mitchell GH, Blackman MJ (2005) Molecular identification of a malaria merozoite surface sheddase" PLoS Pathog 1(3) 241-251
- ^ Child MA, Harris PK, Collins CR, Withers-Martinez C, Yeoh S, Blackman MJ (2013) Molecular determinants for subcellular trafficking of the malarial sheddase PfSUB2. Traffic doi:10.1111/tra.12092
- ^ Alam A, Bhatnagar RK, Relan U, Mukherjee P, Chauhan VS (2013) Proteolytic activity of Plasmodium falciparum subtilisin-like protease 3 on parasite profilin, a multifunctional protein. Mol Biochem Parasitol pii: S0166-6851(13)00137-0. doi:10.1016/j.molbiopara.2013.09.006
- ^ a b Baker RP, Wijetilaka R, Urban S (2006) Two Plasmodium rhomboid proteases preferentially cleave different adhesins implicated in all invasive stages of malaria" PLoS Pathog 2(10) e113
- ^ Singh S, Plassmeyer M, Gaur D, Miller LH (2007) Mononeme: a new secretory organelle in Plasmodium falciparum merozoites identified by localization of rhomboid-1 protease" Proc Natl Acad Sci USA 104(50) 20043-20048
- ^ Deligianni E, Morgan RN, Bertuccini L, Kooij TW, Laforge A, Nahar C, Poulakakis N, Schüler H, Louis C, Matuschewski K, Siden-Kiamos I (2011) Critical role for a stage-specific actin in male exflagellation of the malaria parasite. Cell Microbiol doi:10.1111/j.1462-5822.2011.01652.x
- ^ Wong W, Skau CT, Marapana DS, Hanssen E, Taylor NL, Riglar DT, Zuccala ES, Angrisano F, Lewis H, Catimel B, Clarke OB, Kershaw NJ, Perugini MA, Kovar DR, Gulbis JM, Baum J (2011). "Minimal requirements for actin filament disassembly revealed by structural analysis of malaria parasite actin-depolymerizing factor 1". Proc Natl Acad Sci USA. 108 (24): 9869–74. doi:10.1073/pnas.1018927108. PMC 3116436. PMID 21628589.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Singh BK, Sattler JM, Chatterjee M, Huttu J, Schüler H, Kursula I (2011). "Crystal structures explain functional differences in the two actin depolymerization factors of the malaria parasite". J Biol Chem. 286 (32): 28256–28264. doi:10.1074/jbc.M111.211730. PMC 3151070. PMID 21832095.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Skillman KM, Diraviyam K, Khan A, Tang K, Sept D, Sibley LD (2011). Striepen, Boris (ed.). "Evolutionarily divergent, unstable filamentous actin is essential for gliding motility in apicomplexan parasites". PLoS Pathog. 7 (10): e1002280. doi:10.1371/journal.ppat.1002280.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Angrisano F, Delves MJ, Sturm A, Mollard V, McFadden GI, Sinden RE, Baum J (2011). "A GFP-actin reporter line to explore microfilament dynamics across the malaria parasite lifecycle". Mol Biochem Parasitol.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Ignatev, A; Bhargav, SP; Vahokoski, J; Kursula, P; Kursula, I (2012). Frischknecht, Friedrich (ed.). "The lasso segment is required for functional dimerization of the Plasmodium formin 1 FH2 domain". PLoS ONE. 7 (3): e33586. doi:10.1371/journal.pone.0033586.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Makkonen M, Bertling E, Chebotareva N, Baum J, Lappalainen P (2012) Mammalian and malaria parasite cyclase-associated proteins catalyze nucleotide exchange on G-actin Through a conserved mechanism. J Biol Chem
- ^ Ponts N, Saraf A, Chung DW, Harris A, Prudhomme J, Washburn MP, Florens L, Le Roch KG (2011). "Unraveling the human malaria parasite's ubiquitome". J Biol Chem. 286 (46): 40320–30. doi:10.1074/jbc.M111.238790. PMC 3220526. PMID 21930698.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Certad G, Abrahem A, Georges E (1999) Cloning and partial characterization of the proteasome S4 ATPase from Plasmodium falciparum. Exp Parasitol 93(3) 123-131
- ^ Reiter K, Mukhopadhyay D, Zhang H, Boucher LE, Kumar N, Bosch J, Matunis MJ (2013) Identification of biochemically distinct properties of the SUMO conjugation pathway in Plasmodium falciparum. J Biol Chem
- ^ Hunt P, Afonso A, Creasey A, Culleton R, Sidhu AB, Logan J, Valderramos SG, McNae I, Cheesman S, do Rosario V, Carter R, Fidock DA, Cravo P (2007) Gene encoding a deubiquitinating enzyme is mutated in artesunate- and chloroquine-resistant rodent malaria parasites" Mol Microbiol 65(1) 27-40
- ^ Agrawal S, Chung DW, Ponts N, van Dooren GG, Prudhomme J, Brooks CF, Rodrigues EM, Tan JC, Ferdig MT, Striepen B, Le Roch KG (20130 An apicoplast localized ubiquitylation system is required for the import of nuclear-encoded plastid roteins" PLoS Pathog 9(6) e1003426. doi:10.1371/journal.ppat.1003426
- ^ Botha M, Chiang AN, Needham PG, Stephens LL, Hoppe HC, Külzer S, Przyborski JM, Lingelbach K, Wipf P, Brodsky JL, Shonhai A, Blatch GL (2010). "Plasmodium falciparum encodes a single cytosolic type I Hsp40 that functionally interacts with Hsp70 and is upregulated by heat shock". Cell Stress Chaperones.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Montagna GN, Buscaglia CA, Munter S, Goosmann C, Frischknecht F, Brinkmann V, Matuschewski K (2011) Critical role for heat shock protein 20 (HSP20) in migration of malarial sporozoites. J Biol Chem
- ^ Morahan BJ, Strobel C, Hasan U, Czesny B, Mantel PY, Marti M, Eksi S, Williamson KC (2011). "Functional analysis of the exported type IV HSP40 protein PfGECO in P. falciparum gametocytes". Eukaryotic Cell. 10 (11): 1492. doi:10.1128/EC.05155-11.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Acharya P, Chaubey S, Grover M, Tatu U (2012) An exported heat shock protein 40 associates with pathogenesis-related knobs in Plasmodium falciparum infected erythrocytes" PLoS One 7(9) e44605
- ^ Hatherley R, Blatch GL, Bishop OT (2013) Plasmodium falciparum Hsp70-x: a heat shock protein at the host-parasite interface. J Biomol Struct Dyn
- ^ Gitau GW, Mandal P, Blatch GL, Przyborski J, Shonhai A (2011) Characterisation of the Plasmodium falciparum Hsp70-Hsp90 organising protein (PfHop). Cell Stress Chaperones
- ^ Grover M, Chaubey S, Ranade S, Tatu U (2013) Identification of an exported heat shock protein 70 in Plasmodium falciparum. Parasite 20:2
- ^ Chua CS, Low H, Lehming N, Sim TS (2011). "Molecular analysis of Plasmodium falciparum co-chaperone Aha1 supports its interaction with and regulation of Hsp90 in the malaria parasite". Int J Biochem Cell Biol.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Muralidharan V, Oksman A, Pal P, Lindquist S, Goldberg DE (2012) Plasmodium falciparum heat shock protein 110 stabilizes the asparagine repeat-rich parasite proteome during malarial fevers. Nat Commun 3:1310. doi:10.1038/ncomms2306
- ^ Vitlin Gruber A, Nisemblat S, Zizelski G, Parnas A, Dzikowski R, Azem A, Weiss C (2013) P. falciparum cpn20 Is a bona fide co-chaperonin that can replace GroES in E. coli" PLoS One 8(1) e53909. doi:10.1371/journal.pone.0053909
- ^ Atamna H, Ginsburg H (1997) The malaria parasite supplies glutathione to its host cell - investigation of glutathione transport and metabolism in human erythrocytes infected with Plasmodium falciparum" Eur J Biochem 250(3) 670-679
- ^ Patzewitz EM, Wong EH, Müller S (2011) Dissecting the role of glutathione biosynthesis in Plasmodium falciparum. Mol Microbiol doi:10.1111/j.1365-2958.2011.07933.x
- ^ a b Jortzik E, Becker K (2012) Thioredoxin and glutathione systems in Plasmodium falciparum. Int J Med Microbiol
- ^ Barrand MA, Winterberg M, Ng F, Nguyen M, Kirk K, Hladky SB (2012) Glutathione export from human erythrocytes and Plasmodium falciparum malaria parasites. Biochem J
- ^ Patzewitz EM, Salcedo-Sora JE, Wong EH, Sethia S, Stocks PA, Maughan SC, Murray JA, Krishna S, Bray PG, Ward SA, Müller S (2012) Glutathione transport: A new role for PfCRT in chloroquine resistance. Antioxid Redox Signal
- ^ Kimura R, Komaki-Yasuda K, Kawazu SI, Kano S (2012) 2-Cys peroxiredoxin of Plasmodium falciparum is involved in resistance to heat stress of the parasite. Parasitol Int pii: S1383-5769(12)00157-2. doi:10.1016/j.parint.2012.11.005
- ^ Usui M, Masuda-Suganuma H, Fukumoto S, Angeles JM, Inoue N, Kawazu SI (2012) Expression profiles of peroxiredoxins in liver stage of the rodent malaria parasite Plasmodium berghei. Parasitol Int pii: S1383-5769(12)00159-6. doi:10.1016/j.parint.2012.11.007
- ^ Hakimi H, Asada M, Angeles JM, Kawai S, Inoue N, Kawazu SI (2012) Plasmodium vivax and P. knowlesi: cloning, expression and functional analysis of 1-Cys peroxiredoxin. Exp Parasitol pii: S0014-4894(12)00329-3. doi:10.1016/j.exppara.2012.10.018
- ^ Djuika CF, Fiedler S, Schnölzer M, Sanchez C, Lanzer M, Deponte M (2013) Plasmodium falciparum antioxidant protein as a model enzyme for a special class of glutaredoxin/glutathione-dependent peroxiredoxins. Biochim Biophys Acta pii: S0304-4165(13)00148-7. doi:10.1016/j.bbagen.2013.04.020
- ^ Fritz-Wolf K, Jortzik E, Stumpf M, Preuss J, Iozef R, Rahlfs S, Becker K (2013) Crystal structure of the Plasmodium falciparum thioredoxin reductase-thioredoxin complex.J Mol Biol pii: S0022-2836(13)00432-4. doi:10.1016/j.jmb.2013.06.037
- ^ Ginsburg H, Golenser J (2003) Glutathione is involved in the antimalarial action of chloroquine and its modulation affects drug sensitivity of human and murine species of Plasmodium. Redox Rep 8(5) 276-279
- ^ Putonti C, Quach B, Kooistra RA, Kanzok SM (2012) The evolution and putative function of phosducin-like proteins in the malaria parasite Plasmodium. Infect Genet Evol pii: S1567-1348(12)00294-8. doi:10.1016/j.meegid.2012.08.023
- ^ Boysen KE, Matuschewski K (2011). "Arrested oocyst maturation in Plasmodium parasites lacking type II NADH:ubiquinone dehydrogenase". J Biol Chem. 286 (37): 32661–71. doi:10.1074/jbc.M111.269399. PMC 3173203. PMID 21771793.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Knöckel J, Müller IB, Butzloff S, Bergmann B, Walter RD, Wrenger C (2012). "The antioxidative effect of de novo generated vitamin B6 in Plasmodium falciparum validated by protein interference". Biochem J. 443 (2): 397–405. doi:10.1042/BJ20111542. PMID 22242896.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Matthews K, Kalanon M, Chisholm SA, Sturm A, Goodman CD, Dixon MW, Sanders PR, Nebl T, Fraser F, Haase S, McFadden GI, Gilson PR, Crabb BS, de Koning-Ward TF (2013) The Plasmodium translocon of exported proteins (PTEX) component thioredoxin-2 is important for maintaining normal blood-stage growth. Mol Microbiol doi:10.1111/mmi.12334
- ^ Sousa Silva M, Ferreira AE, Gomes R, Tomás AM, Ponces Freire A, Cordeiro C (2012) The glyoxalase pathway in protozoan parasites. Int J Med Microbiol 302(4-5) 225-229 doi:10.1016/j.ijmm.2012.07.005
- ^ Wasserman M, Vernot JP, Mendoza PM (1990) Role of calcium and erythrocyte cytoskeleton phosphorylation in the invasion of Plasmodium falciparum. Parasitol Res 76(8) 681-688
- ^ Tanabe K (1990) Ion metabolism in malaria-infected erythrocytes. Blood Cells 16(2-3) 437-449
- ^ Enomoto M, Kawazu S, Kawai S, Furuyama W, Ikegami T, Watanabe J, Mikoshiba K (2012) Blockage of spontaneous Ca(2+) oscillation causes cell death in intraerythrocitic Plasmodium falciparum" PLoS One 7(7) e39499.
- ^ Stritzke C, Nalaskowski MM, Fanick W, Lin H, Mayr GW (2012) A Plasmodium multi-domain protein possesses multiple inositol phosphate kinase activities. Mol Biochem Parasitol pii: S0166-6851(12)00249-6. doi:10.1016/j.molbiopara.2012.10.005
- ^ Polson HE, Blackman MJ (2005) A role for poly(dA)poly(dT) tracts in directing activity of the Plasmodium falciparum calmodulin gene promoter. Mol Biochem Parasitol 141(2) 179-189
- ^ Raabe A, Berry L, Sollelis L, Cerdan R, Tawk L, Vial HJ, Billker O, Wengelnik K (2011) Genetic and transcriptional analysis of phosphoinositide-specific phospholipase C in Plasmodium Exp Parasitol
- ^ Clinch K, Crump DR, Evans GB, Hazleton KZ, Mason JM, Schramm VL, Tyler PC (2013) cyclic phosph(on)ate inhibitors of Plasmodium falciparum hypoxanthine-guanine-xanthine phosphoribosyltransferase. Bioorg Med Chem pii: S0968-0896(13)00140-5. doi:10.1016/j.bmc.2013.02.016
- ^ Spalding MD, Allary M, Gallagher JR, Prigge ST (2010). "Validation of a modified method for Bxb1 mycobacteriophage integrase-mediated recombination in Plasmodium falciparum by localization of the H-protein of the glycine cleavage complex to the mitochondrion. Mol. Biochem". Parasitol.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Pornthanakasem W, Kongkasuriyachai D, Uthaipibull C, Yuthavong Y, Leartsakulpanich U (2012) Plasmodium serine hydroxymethyltransferase: indispensability and display of distinct localization. Malar J 11(1) 387
- ^ Suthar MK, Verma A, Doharey PK, Singh SV, Saxena JK (2013) Single tryptophan of disordered loop from Plasmodium falciparum purine nucleoside phosphorylase: Involvement in catalysis and microenvironment. Appl Biochem Biotechnol
- ^ Takashima Y, Mizohata E, Tokuoka K, Krungkrai SR, Kusakari Y, Konishi S, Satoh A, Matsumura H, Krungkrai J, Horii T, Inoue T (2012) Crystallization and preliminary X-ray diffraction analysis of orotate phosphoribosyltransferase from the human malaria parasite Plasmodium falciparum. Acta Crystallogr Sect F Struct Biol Cryst Commun 68(2) 244-246
- ^ Guler JL, Freeman DL, Ahyong V, Patrapuvich R, White J, Gujjar R, Phillips MA, Derisi J, Rathod PK (2013) Asexual populations of the human malaria parasite, Plasmodium falciparum, use a two-step genomic strategy to acquire accurate, beneficial DNA amplifications" PLoS Pathog 9(5) e1003375. doi:10.1371/journal.ppat.1003375
- ^ Hoeijmakers WA, Flueck C, Françoijs KJ, Smits AH, Wetzel J, Volz JC, Cowman AF, Voss T, Stunnenberg HG, Bártfai R (2012) Plasmodium falciparum centromeres display a unique epigenetic makeup and cluster prior to and during schizogony. Cell Microbiol doi:10.1111/j.1462-5822.2012.01803.x
- ^ Schoenfeld TW, Murugapiran S, Dodsworth JA, Floyd S, Lodes M, Mead DA, Hedlund BP (2013) Lateral gene transfer of Family A DNA polymerases between thermophilic viruses, Aquificae, and Apicomplexa. Mol Biol Evol
- ^ Ghorbal M, Scheidig-Benatar C, Bouizem S, Thomas C, Paisley G, Faltermeier C, Liu M, Scherf A, Lopez-Rubio JJ, Gopaul DN (2012) Initial characterization of the Pf-int recombinase from the malaria parasite Plasmodium falciparum" PLoS One 7(10) e46507. doi:10.1371/journal.pone.0046507
- ^ Ansari A, Tuteja R (2012) Genome wide comparative comprehensive analysis of Plasmodium falciparum MCM family with human host. Commun Integr Biol 5(6) 607-615
- ^ Flueck C, Bartfai R, Niederwieser I, Witmer K, Alako BT, Moes S, Bozdech Z, Jenoe P, Stunnenberg HG, Voss TS (2010) A major role for the Plasmodium falciparum ApiAP2 protein PfSIP2 in chromosome end biology" PLoS Pathog 6(2) e1000784. doi:10.1371/journal.ppat.1000784
- ^ Chêne A, Vembar SS, Rivière L, Lopez-Rubio JJ, Claes A, Siegel TN, Sakamoto H, Scheidig-Benatar C, Hernandez-Rivas R, Scherf A (2011). "PfAlbas constitute a new eukaryotic DNA/RNA-binding protein family in malaria parasites". Nucleic Acids Res.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Antony E, Weiland EA, Korolev S, Lohman TM (2012) Plasmodium falciparum SSB tetramer wraps single stranded DNA with similar topology but opposite polarity to E. coli SSB. J Mol Biol
- ^ Antony E, Kozlov AG, Nguyen B, Lohman TM (2012) Plasmodium falciparum SSB tetramer binds single stranded DNA only in a fully wrapped mode. J Mol Biol
- ^ a b Gopalakrishnan AM, Kumar N (2013) Opposing roles for two molecular forms of replication protein A in Rad51-Rad54-mediated DNA recombination in Plasmodium falciparum. MBio 4(3) pii: e00252-13. doi:10.1128/mBio.00252-13
- ^ Pace T, Olivieri A, Sanchez M, Albanesi V, Picci L, Siden Kiamos I, Janse CJ, Waters AP, Pizzi E, Ponzi M (2006) Set regulation in asexual and sexual Plasmodium parasites reveals a novel mechanism of stage-specific expression" Mol Microbiol 60(4) 870-882
- ^ Choi SW, Keyes MK, Horrocks P (2006) LC/ESI-MS demonstrates the absence of 5-methyl-2′-deoxycytosine in Plasmodium falciparum genomic DNA. Mol Biochem Parasitol 150:350–352
- ^ Tarique M, Ahmad M, Ansari A, Tuteja R (2013) Plasmodium falciparum DOZI, an RNA helicase interacts with eIF4E. Gene pii: S0378-1119(13)00332-6. doi:10.1016/j.gene.2013.03.063
- ^ Komaki-Yasuda K, Okuwaki M, Nagata K, Kawazu S, Kano S (2013) Identification of a novel and unique transcription factor in the intraerythrocytic stage of Plasmodium falciparum" PLoS One 8(9) e74701 doi:10.1371/journal.pone.0074701
- ^ Tarique M, Satsangi AT, Ahmad M, Singh S, Tuteja R (2011). "Plasmodium falciparum MLH is schizont stage specific endonuclease". Mol Biochem Parasitol.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Siribal S, Weinfeld M, Karimi-Busheri F, Mark Glover JN, Bernstein NK, Aceytuno D, Chavalitshewinkoon-Petmitr P (2011). "Molecular characterization of Plasmodium falciparum putative polynucleotide kinase/phosphatase". Mol Biochem Parasitol. 180 (1): 1–7. doi:10.1016/j.molbiopara.2011.06.007. PMID 21821066.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Ho CK, Shuman S (2001) A yeast-like mRNA capping apparatus in Plasmodium falciparum" Proc Natl Acad Sci USA 98(6) 3050-3035
- ^ Eshar S, Allemand E, Sebag A, Glaser F, Muchardt C, Mandel-Gutfreund Y, Karni R, Dzikowski R (2012) A novel Plasmodium falciparum SR protein is an alternative splicing factor required for the parasites' proliferation in human erythrocytes. Nucleic Acids Res
- ^ Singh PK, Kanodia S, Dandin CJ, Vijayraghavan U, Malhotra P (2012) Plasmodium falciparum Prp16 homologue and its role in splicing. Biochim Biophys Acta pii: S1874-9399(12)00157-5. doi:10.1016/j.bbagrm.2012.08.014
- ^ Kishore SP, Perkins SL, Templeton TJ, Deitsch KW (2009) An unusual recent expansion of the C-terminal domain of RNA polymerase II in primate malaria parasites features a motif otherwise found only in mammalian polymerases" J Mol Evol 68(6) 706-14. doi:10.1007/s00239-009-9245-2 PMID 19449052
- ^ Kaushik A, Subramaniam S, Gupta D (2013) In silico characterization and molecular dynamics simulation of Pfcyc-1, a cyclin homolog of Plasmodium falciparum. J Biomol Struct Dyn
- ^ Varunan SM, Tripathi J, Bhattacharyya S, Suhane T, Bhattacharyya MK () Plasmodium falciparum origin recognition complex subunit 1 (PfOrc1) functionally complements Δsir3 mutant of Saccharomyces cerevisiae. doi:10.1016/j.molbiopara.2013.08.004
- ^ Hossain M, Sharma S, Korde R, Kanodia S, Chugh M, Rawat K, Malhotra P (2013) Organization of Plasmodium falciparum spliceosomal core complex and role of arginine methylation in its assembly. Malar J 12(1) 333
- ^ Mehta J, Tuteja R (2011) A novel dual Dbp5/DDX19 homologue from Plasmodium falciparum requires Q motif for activity. Mol Biochem Parasitol 176(1) 58-63 doi:10.1016/j.molbiopara.2010.12.003
- ^ Prakash K, Tuteja R (2010) A novel DEAD box helicase Has1p from Plasmodium falciparum: N-terminal is essential for activity. Parasitol Int 59(2) 271-277 doi:10.1016/j.parint.2010.02.003
- ^ a b Ahmad M, Ansari A, Tarique M, Satsangi AT, Tuteja R (2012) Plasmodium falciparum UvrD helicase translocates in 3' to 5' direction, colocalizes with MLH and modulates Its activity through physical interaction" PLoS One 7(11) e49385. doi:10.1371/journal.pone.0049385 Cite error: The named reference "Ahmad2012" was defined multiple times with different content (see the help page).
- ^ Pradhan A, Tuteja R (2007) Bipolar, dual Plasmodium falciparum helicase 45 expressed in the intraerythrocytic developmental cycle is required for parasite growth" J Mol Biol 373(2) 268-281
- ^ Suntornthiticharoen P, Petmitr S, Chavalitshewinkoon-Petmitr P (2006) Purification and characterization of a novel 3'-5' DNA helicase from Plasmodium falciparum and its sensitivity to anthracycline antibiotics" Parasitology 133(4) 389-398
- ^ Shankar J, Pradhan A, Tuteja R (2008) Isolation and characterization of Plasmodium falciparum UAP56 homolog: evidence for the coupling of RNA binding and splicing activity by site-directed mutations. Arch Biochem Biophys 478(2) 143-53 doi:10.1016/j.abb.2008.07.027
- ^ Ahmad M, Tuteja R (2013) Plasmodium falciparum RuvB1 is an active DNA helicase and translocates in the 5'-3' direction" Gene 515(1) 99-109 doi:10.1016/j.gene.2012.11.020
- ^ Ahmad M, Tuteja R (2013) Plasmodium falciparum RuvB2 translocates in 5'-3' direction, relocalizes during schizont stage and its enzymatic activities are up regulated by RuvB3 of the same complex. Biochim Biophys Acta pii: S1570-9639(13)00368-3. doi:10.1016/j.bbapap.2013.10.010
- ^ Hoeijmakers WA, Salcedo-Amaya AM, Smits AH, Françoijs KJ, Treeck M, Gilberger TW, Stunnenberg HG, Bártfai R (2013) H2A.Z/H2B.Z double-variant nucleosomes inhabit the AT-rich promoter regions of the Plasmodium falciparum genome. Mol Microbiol doi:10.1111/mmi.12151
- ^ Miao J, Fan Q, Cui L, Li J. The malaria parasite Plasmodium falciparum histones: organization, expression, and acetylation" Gene 369:53–65
- ^ a b Trelle MB, Salcedo-Amaya AM, Cohen AM, Stunnenberg HG, Jensen ON. Global histone analysis by mass spectrometry reveals a high content of acetylated lysine residues in the malaria parasite Plasmodium falciparum. J Proteome Res 8:3439–3450
- ^ Flueck C, Bartfai R, Volz J, Niederwieser I, Salcedo-Amaya AM, et al (2009) Plasmodium falciparum heterochromatin protein 1 marks genomic loci linked to phenotypic variation of exported virulence factors" PLoS Pathog 5:e1000569
- ^ Bischoff E, Vaquero C (2010) In silico and biological survey of transcription-associated proteins implicated in the transcriptional machinery during the erythrocytic development of Plasmodium falciparum" BMC Genomics 11:34
- ^ Joshi MB, Lin DT, Chiang PH, Goldman ND, Fujioka H, Aikawa M, Syin C (1999) Molecular cloning and nuclear localization of a histone deacetylase homologue in Plasmodium falciparum. Mol Biochem Parasitol 99(1) 11-19
- ^ Zhu AY, Zhou Y, Khan S, Deitsch K, Hao Q, Lin H (2011). "Plasmodium falciparum Sir2A preferentially hydrolyzes medium and long chain fatty acyl lysine". ACS Chem Biol.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Sierra-Miranda M, Delgadillo DM, Mancio-Silva L, Vargas M, Villegas-Sepulveda N, Martínez-Calvillo S, Scherf A, Hernández-Rivas R (2012) Two long non-coding RNAs generated from subtelomeric regions accumulate in a novel perinuclear compartment in Plasmodium falciparum. Mol Biochem Parasitol
- ^ Harris EY, Ponts N, Le Roch KG, Lonardi S (2011). "Chromatin-driven de novo discovery of DNA binding motifs in the human malaria parasite". BMC Genomics. 12 (1): 601. doi:10.1186/1471-2164-12-601.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Painter HJ, Campbell TL, Llinas M (2011) The Apicomplexan AP2 family: integral factors regulating Plasmodium development. Mol Biochem Parasitol 176: 1–7
- ^ Iwanaga S, Kaneko I, Kato T, Yuda M (2012) Identification of an AP2-family protein that is critical for malaria liver stage development" PLoS One 7(11) e47557. doi:10.1371/journal.pone.0047557
- ^ Lima WR, Moraes M, Alves E, Azevedo MF, Passos DO, Garcia CR (2012) The PfNF-YB transcription factor is a downstream target of melatonin and cAMP signalling in the human malaria parasite Plasmodium falciparum. J Pineal Res doi:10.1111/j.1600-079X.2012.01021.x
- ^ Gissot M, Briquet S, Refour P, Boschet C, Vaquero C (2005). "PfMyb1, a Plasmodium falciparum transcription factor, is required for intra-erythrocytic growth and controls key genes for cell cycle regulation". J Mol Biol. 346 (1): 29–42. doi:10.1016/j.jmb.2004.11.045. PMID 15663925.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Aly, AS; Lindner, SE; MacKellar, DC; Peng, X; Kappe, SH (2011). "SAP1 is a critical post-transcriptional regulator of infectivity in malaria parasite sporozoite stages". Mol Microbiol. 79 (4): 929–939. doi:10.1111/j.1365-2958.2010.07497.x. PMID 21299648.
{{cite journal}}: Unknown parameter|author-separator=ignored (help) - ^ Balu B, Maher SP, Pance A, Chauhan C, Naumov AV, Andrews RM, Ellis PD, Khan SM, Lin JW, Janse CJ, Rayner JC, Adams JH (2011). "CCR4-associated factor-1 coordinates expression of Plasmodium falciparum egress and invasion proteins". Eukaryotic Cell. 10 (9): 1257. doi:10.1128/EC.05099-11.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Bopp SE, Manary MJ, Bright AT, Johnston GL, Dharia NV, Luna FL, McCormack S, Plouffe D, McNamara CW, Walker JR, Fidock DA, Denchi EL, Winzeler EA (2013) Mitotic evolution of Plasmodium falciparum shows a stable core genome but recombination in antigen families" PLoS Genet 9(2) e1003293. doi:10.1371/journal.pgen.1003293
- ^ Atemnkeng VA, Pink M, Schmitz-Spanke S, Wu XJ, Dong LL, Zhao KH, May C, Laufer S, Langer B, Kaiser A (2013) Deoxyhypusine hydroxylase from Plasmodium vivax, the neglected human malaria parasite: molecular cloning, expression and specific inhibition by the 5-LOX inhibitor zileuton" PLoS One 8(3) e58318. doi:10.1371/journal.pone.0058318
- ^ Schwentke A, Krepstakies M, Müller AK, Hammerschmidt C, Motaal B, Bernhard T, Hauber J, Kaiser A (2012) In vitro and in vivo silencing of plasmodial dhs and eIF-5A genes in a putative, non-canonical RNAi-related pathway. BMC Microbiol 12(1) 107
- ^ Johnson RA, McFadden GI, Goodman CD (2011). Langsley, Gordon (ed.). "Characterization of two malaria parasite organelle translation elongation factor g proteins: the likely targets of the anti-malarial fusidic acid". PLoS ONE. 6 (6): e20633. doi:10.1371/journal.pone.0020633. PMC 3112199. PMID 21695207.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Zhang M, Mishra S, Sakthivel R, Rojas M, Ranjan R, Sullivan WJ Jr, Fontoura BM, Ménard R, Dever TE, Nussenzweig V (2012) PK4, a eukaryotic initiation factor 2α(eIF2α) kinase, is essential for the development of the erythrocytic cycle of Plasmodium. Proc Natl Acad Sci USA
- ^ Filisetti D, Theobald-Dietrich A, Mahmoudi N, Rudinger-Thirion J, Candolfi E, Frugier M (2013) Aminoacylation of Plasmodium falciparum tRNAAsn and insights in the synthesis of asparagine repeats. J Biol Chem 2013
- ^ Jackson KE, Pham JS, Kwek M, De Silva NS, Allen SM, Goodman CD, McFadden GI, de Pouplana LR, Ralph SA (2011). "Dual targeting of aminoacyl-tRNA synthetases to the apicoplast and cytosol in Plasmodium falciparum". Int J Parasitol.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Bhatt TK, Khan S, Dwivedi VP, Banday MM, Sharma A, Chandele A, Camacho N, de Pouplana LR, Wu Y, Craig AG, Mikkonen AT, Maier AG, Yogavel M, Sharma A (2011) Malaria parasite tyrosyl-tRNA synthetase secretion triggers pro-inflammatory responses. Nat Commun 2:530. doi:10.1038/ncomms1522
- ^ a b Khan S, Garg A, Camacho N, Van Rooyen J, Kumar Pole A, Belrhali H, Ribas de Pouplana L, Sharma V, Sharma A (2013) Structural analysis of malaria-parasite lysyl-tRNA synthetase provides a platform for drug development. Acta Crystallogr D Biol Crystallogr 69(5) 785-795 doi:10.1107/S0907444913001923 Cite error: The named reference "Khan2013" was defined multiple times with different content (see the help page).
- ^ Jones ML, Collins MO, Goulding D, Choudhary JS, Rayner JC (2012) Analysis of protein palmitoylation reveals a pervasive role in Plasmodium development and pathogenesis. Cell Host Microbe 12(2) 246-258
- ^ Rackham MD, Brannigan JA, Moss DK, Yu Z, Wilkinson AJ, Holder AA, Tate EW, Leatherbarrow RJ (2012) Discovery of novel and ligand-efficient inhibitors of Plasmodium falciparum and Plasmodium vivax N-Myristoyltransferase. J Med Chem
- ^ Hempelmann E. (2007). "Hemozoin biocrystallization in Plasmodium falciparum and the antimalarial activity of crystallization inhibitors". Parasitol Research. 100 (4): 671–676. doi:10.1007/s00436-006-0313-x. PMID 17111179.[dead link]
- ^ Bonday, Z.Q.; Dhanasekaran, S; Rangarajan, PN; Padmanaban, G (2002). "Import of host delta-aminolevulinate dehydratase into the malarial parasite: Identification of a new drug target". Nature Medicine. 6 (8): 898–903. doi:10.1038/78659. PMID 10932227.
- ^ Nagaraj VA, Sundaram B, Varadarajan NM, Subramani PA, Kalappa DM, Ghosh SK, Padmanaban G (2013) Malaria parasite-synthesized heme is essential in the mosquito and liver stages and complements host heme in the blood stages of infection. PloS Pathog (8) e1003522. doi:10.1371/journal.ppat.1003522
- ^ Bohle DS, Dodd EL, Stephens PW (2012) Structure of malaria pigment and related propanoate-linked metalloporphyrin dimers. Chem Biodivers 9(9) 1891-1902 doi:10.1002/cbdv.201200033
- ^ Cortopassi WA, Oliveira AA, Guimaraes AP, Renno MN, Krettli AU, Franca TC (2011) Docking studies on the binding of quinoline derivatives and hematin to Plasmodium falciparum lactate dehydrogenase. J Biomol Struct Dyn 29(1) 207-218
- ^ Chugh M, Sundararaman V, Kumar S, Reddy VS, Siddiqui WA, Stuart KD, Malhotra P (2013) Protein complex directs hemoglobin-to-hemozoin formation in Plasmodium falciparum. Proc Natl Acad Sci USA
- ^ Nakatani K, Ishikawa H, Aono S, Mizutani Y (2013) Heme-binding properties of heme detoxification protein from Plasmodium falciparum. Biochem Biophys Res Commun pii: S0006-291X(13)01458-7. doi:10.1016/j.bbrc.2013.08.100
- ^ Sigala PA, Crowley JR, Hsieh S, Henderson JP, Goldberg DE (2012) Direct tests of enzymatic heme degradation by the malaria parasite Plasmodium falciparum. J Biol Chem
- ^ Klonis N, Crespo-Ortiz MP, Bottova I, Abu-Bakar N, Kenny S, Rosenthal PJ, Tilley L (2011). "Artemisinin activity against Plasmodium falciparum requires hemoglobin uptake and digestion". Proc Natl Acad Sci U S A. 108 (28): 11405–10. doi:10.1073/pnas.1104063108. PMC 3136263. PMID 21709259.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b Combrinck JM, Mabotha TE, Ncokazi KK, Ambele MA, Taylor D, Smith PJ, Hoppe HC, Egan TJ (2012) Insights into the role of heme in the mechanism of action of ntimalarials. ACS Chem Biol
- ^ Prato M, D'Alessandro S, Van den Steen PE, Opdenakker G, Arese P, Taramelli D, Basilico N (2011) Natural haemozoin modulates matrix metalloproteinases and induces morphological changes in human microvascular endothelium. Cell Microbiol. doi:10.1111/j.1462-5822.2011.01620.x
- ^ Deroost K, Tyberghein A, Lays N, Noppen S, Schwarzer E, Vanstreels E, Komuta M, Prato M, Lin JW, Pamplona A, Janse CJ, Arese P, Roskams T, Daelemans D, Opdenakker G, Van den Steen PE (2013) Hemozoin induces lung inflammation and correlates with malaria-associated acute respiratory distress syndrome. Am J Respir Cell Mol Biol
- ^ a b Polimeni M, Valente E, Aldieri E, Khadjavi A, Giribaldi G, Prato M (2013) Role of 15-hydroxyeicosatetraenoic acid in hemozoin-induced lysozyme release from human adherent monocytes. Biofactors doi:10.1002/biof.1071 Cite error: The named reference "Polimeni2013" was defined multiple times with different content (see the help page).
- ^ Tyberghein A, Deroost K, Schwarzer E, Arese P, Van den Steen PE (2013) Immunopathological effects of malaria pigment or hemozoin and other crystals. Biofactors doi:10.1002/biof.1119
- ^ Thawani N, Tam M, Bellemare MJ, Bohle DS, Olivier M, de Souza JB, Stevenson MM (2013) Haemozoin inhibts erythropoetin induced proliferation of erythroid precursors. J Infect Dis
- ^ Lian LY, Al-Helal M, Roslaini AM, Fisher N, Bray PG, Ward SA, Biagini GA (2009) Glycerol: an unexpected major metabolite of energy metabolism by the human malaria parasite. Malar J 8:38. doi:10.1186/1475-2875-8-38
- ^ Sanz S, Bandini G, Ospina D, Bernabeu M, Mariño K, Fernández-Becerra C, Izquierdo L (2013) Biosynthesis of GDP-fucose and other sugar nucleotides in the blood-stages of Plasmodium falciparum. J Biol Chem
- ^ Macrae JI, Dixon MW, Dearnley MK, Chua HH, Chambers JM, Kenny S, Bottova I, Tilley L, McConville MJ (2013) Mitochondrial metabolism of sexual and asexual blood stages of the malaria parasite Plasmodium falciparum. BMC Biol 11(1) 67
- ^ Tjhin ET, Staines HM, van Schalkwyk DA, Krishna S, Saliba KJ (2013) Studies with the Plasmodium falciparum hexokinase reveal that PfHT limits the rate of glucose entry into glycolysis. FEBS Lett pii: S0014-5793(13)00618-2. doi:10.1016/j.febslet.2013.07.052
- ^ Naidoo K, Coetzer TL (2013) Reduced intra-erythrocytic growth of glycerol kinase knockout Plasmodium falciparum parasites during in vitro blood stage development. Biochim Biophys Acta pii: S0304-4165(13)00350-4. doi:10.1016/j.bbagen.2013.08.006
- ^ Bobenchik AM, Witola WH, Augagneur Y, Nic Lochlainn L, Garg A, Pachikara N, Choi JY, Zhao YO, Usmani-Brown S, Lee A, Adjalley SH, Samanta S, Fidock DA, Voelker DR, Fikrig E, Ben Mamoun C (2013) Plasmodium falciparum phosphoethanolamine methyltransferase is essential for malaria transmission. Proc Natl Acad Sci USA
- ^ Sen P, Vial HJ, Radulescu O (2013) Kinetic modelling of phospholipid synthesis in Plasmodium knowlesi unravels crucial steps and relative importance of multiple pathways. BMC Syst Biol 7(1):123
- ^ Maheshwari S, Lavigne M, Contet A, Alberge B, Pihan E, Kocken C, Wengelnik K, Douguet D, Vial H, Cerdan R (2012) Biochemical characterization of Plasmodium falciparum CTP:phosphoethanolamine cytidylyltransferase shows that only one of the two cytidylyltransferase domains is active. Biochem J
- ^ Choi JY, Augagneur Y, Ben Mamoun C, Voelker DR (2011). "Identification of a gene encoding Plasmodium knowlesi phosphatidylserine decarboxylase by genetic complementation in yeast, and characterization of in vitro maturation of the encoded enzyme". J Biol Chem.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Maity K, Venkata BS, Kapoor N, Surolia N, Surolia A, Suguna K (2011) Structural basis for the functional and inhibitory mechanisms of β-hydroxyacyl-acyl carrier protein dehydratase (FabZ) of Plasmodium falciparum. J Struct Biol
- ^ a b c Storm J, Müller S (2012) lipoic acid metabolism of Plasmodium - A suitable drug target. Curr Pharm Des
- ^ Lauinger IL, Vivas L, Perozzo R, Stairiker C, Tarun A, Zloh M, Zhang X, Xu H, Tonge PJ, Franzblau SG, Pham DH, Esguerra CV, Crawford AD, Maes L, Tasdemir D (2013) Potential of lichen secondary metabolites against Plasmodium liver stage parasites with FAS-II as the potential target. J Nat Prod 76(6) 1064-1070
- ^ Umeda T, Tanaka N, Kusakabe Y, Nakanishi M, Kitade Y, Nakamura KT (2011) Molecular basis of fosmidomycin's action on the human malaria parasite Plasmodium falciparum. Sci Rep 1:9
- ^ Howe R, Kelly M, Jimah J, Hodge D, Odom AR (2012) Isoprenoid biosynthesis inhibition disrupts Rab5 localization and food vacuolar integrity in Plasmodium falciparum. Eukaryot Cell
- ^ Liu YL, Guerra F, Wang K, Wang W, Li J, Huang C, Zhu W, Houlihan K, Li Z, Zhang Y, Nair SK, Oldfield E (2012) Structure, function and inhibition of the two- and three-domain 4Fe-4S IspG proteins. Proc Natl Acad Sci USA
- ^ Pratt S, Wansadhipathi-Kannangara NK, Bruce CR, Mina JG, Shams-Eldin H, Casas J, Hanada K, Schwarz RT, Sonda S, Denny PW (2012) Sphingolipid synthesis and scavenging in the intracellular apicomplexan parasite, Toxoplasma gondii. Mol Biochem Parasitol pii: S0166-6851(12)00283-6. doi:10.1016/j.molbiopara.2012.11.007
- ^ Lee SG, Kim Y, Alpert TD, Nagata A, Jez JM (2011). "Structure and reaction mechanism of phosphoethanolamine methyltransferase from the malaria parasite Plasmodium falciparum: An anti-parasitic drug target". J Biol Chem.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Walker DM, Mahfooz N, Kemme KA, Patel VC, Spangler M, Drew ME (2013) Plasmodium falciparum erythrocytic stage parasites require the putative autophagy protein PfAtg7 for normal growth" PLoS One 8(6) e67047
- ^ Ginsburg, H; Gorodetsky, R; Krugliak, M (1986). "The status of zinc in malaria (Plasmodium falciparum) infected human red blood cells: stage dependent accumulation, compartmentation and effect of dipicolinate". Biochim Biophys Acta. 886 (3): 337–344. doi:10.1016/0167-4889(86)90168-0. PMID 3518809.
- ^ Niles JC (2012) Malarial parasites accumulate labile zinc pools. Chem Biol 19(6) 660-661
- ^ Asahi H, Tolba ME, Tanabe M, Ohmae H (2013) Molecular factors that are associated with early developmental arrest of intraerythrocytic Plasmodium falciparum. Can J Microbiol 59(7) 485-93. doi:10.1139/cjm-2013-0166
- ^ Mawson AR (2013) The pathogenesis of malaria: a new perspective. Pathog Glob Health 107(3) 122-129
- ^ Williams M, Sprenger J, Human E, Al-Karadaghi S, Persson L, Louw AI, Birkholtz LM (2011). "Biochemical characterisation and novel classification of monofunctional S-adenosylmethionine decarboxylase of Plasmodium falciparum". Mol Biochem Parasitol. 180 (1): 17–26. doi:10.1016/j.molbiopara.2011.07.004. PMID 21803076.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Jortzik E, Mailu BM, Preuss J, Fischer M, Bode L, Rahlfs S, Becker K (2011). "Glucose 6-phosphate dehydrogenase 6-phosphogluconolactonase: a unique bifunctional enzyme from Plasmodium falciparum". Biochem J. 436 (3): 641–50. doi:10.1042/BJ20110170. PMID 21443518.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Stover, NA; Dixon, TA; Cavalcanti, AR (2011). Moreno, Silvia N (ed.). "Multiple independent fusions of glucose-6-phosphate dehydrogenase with enzymes in the pentose phosphate pathway". PLoS ONE. 6 (8): e22269. doi:10.1371/journal.pone.0022269.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Samanta M, Murthy MR, Balaram H, Balaram P (2011) Revisiting the mechanism of the triose-phosphate isomerase reaction: The role of the fully conserved glutamic acid 97 Residue. Chembiochem doi:10.1002/cbic.201100116
- ^ Sussmann RA, Angeli CB, Peres VJ, Kimura EA, Katzin AM (2011). "Intraerythrocytic stages of Plasmodium falciparum biosynthesize vitamin E". FEBS Lett. 585 (24): 3985–91. doi:10.1016/j.febslet.2011.11.005. PMID 22085796.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Guédez G, Hipp K, Windeisen V, Derrer B, Gengenbacher M, Böttcher B, Sinning I, Kappes B, Tews I; et al. (2012). "Assembly of the eukaryotic PLP-synthase complex from Plasmodium and activation of the Pdx1 enzyme". Structure. 20 (1): 172–184. doi:10.1016/j.str.2011.11.015. PMID 22244765.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Tapas S, Kumar A, Dhindwal S, Preeti, Kumar P (2011). "Structural analysis of chorismate synthase from Plasmodium falciparum: A novel target for antimalaria drug discovery". Int J Biol Macromol. 49 (4): 767–77. doi:10.1016/j.ijbiomac.2011.07.011. PMID 21801743.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Zocher K, Fritz-Wolf K, Kehr S, Fischer M, Rahlfs S, Becker K (2012) Biochemical and structural characterization of Plasmodium falciparum glutamate dehydrogenase 2. Mol Biochem Parasitol
- ^ Salazar E, Bank EM, Ramsey N, Hess KC, Deitsch KW, Levin LR, Buck J (2012) Characterization of Plasmodium falciparum adenylyl cyclase-β and its role in erythrocytic stage parasites" PLoS One 7(6) e39769.
- ^ Osman W, Endo S, Oh-Hashi K, Kitamura Y, Kitade Y (2012) Molecular characterization and mutational analysis of recombinant diadenosine 5',5″-P1,P4-tetraphosphate hydrolase from Plasmodium falciparum. Biol Pharm Bull 35(7) 1191-1196
- ^ Denloye T, Dalal S, Klemba M (2012) Characterization of a glycerophosphodiesterase with an unusual tripartite distribution and an important role in the asexual blood stages of Plasmodium falciparum. Mol Biochem Parasitol pii: S0166-6851(12)00222-8. doi:10.1016/j.molbiopara.2012.09.004
- ^ Eichhorn T, Winter D, Büchele B, Dirdjaja N, Frank M, Lehmann WD, Mertens R, Krauth-Siegel RL, Simmet T, Granzin J, Efferth T (2012) Molecular interaction of artemisinin with translationally controlled tumor protein (TCTP) of Plasmodium falciparum. Biochem Pharmacol pii: S0006-2952(12)00678-8. doi:10.1016/j.bcp.2012.10.006
- ^ Jordão FM, Gabriel HB, Alves JM, Angeli CB, Bifano TD, Breda A, de Azevedo MF, Basso LA, Wunderlich G, Kimura EA, Katzin AM (2013) Cloning and characterization of bifunctional enzyme farnesyl iphosphate/geranylgeranyl diphosphate synthase from Plasmodium falciparum. Malar J 12(1) 184
- ^ Chan XW, Wrenger C, Stahl K, Bergmann B, Winterberg M, Müller IB, Saliba KJ (2013) Chemical and genetic validation of thiamine utilization as an antimalarial drug target. Nat Commun 4:2060. doi:10.1038/ncomms3060
- ^ Ojha PK, Roy K (2013) First report on exploring structural requirements of alpha and beta thymidine analogs for PfTMPK inhibitory activity using in silico studies. Biosystems pii: S0303-2647(13)00165-2 doi:10.1016/j.biosystems.2013.07.005
- ^ Duffy MF, Byrne TJ, Carret C, Ivens A, Brown GV (2009) Ectopic recombination of a malaria var gene during mitosis associated with an altered var switch rate" J Mol Biol 389: 453–469 doi:10.1016/j.jmb.2009.04.032
- ^ Rask TS, Hansen DA, Theander TG, Gorm PA, Lavstsen T (2010) Plasmodium falciparum erythrocyte membrane protein 1 diversity in seven genomes--divide and conquer. PLOS Comput Biol 6: e1000933.
- ^ a b Chen Q, Heddini A, Barragan A, Fernandez V, Pearce SF, et al. (2000) The semiconserved head structure of Plasmodium falciparum erythrocyte membrane protein 1 mediates binding to multiple independent host receptors" J Exp Med 192: 1–10. doi:10.1084/jem.192.1.1 Cite error: The named reference "Chen2000" was defined multiple times with different content (see the help page).
- ^ Smith JD, Subramanian G, Gamain B, Baruch DI, Miller LH (2000) Classification of adhesive domains in the Plasmodium falciparum erythrocyte membrane protein 1 family. Mol Biochem Parasitol 110: 293–310. doi:10.1016/S0166-6851(00)00279-6
- ^ Robinson BA, Welch TL, Smith JD (2003) Widespread functional specialization of Plasmodium falciparum erythrocyte membrane protein 1 family members to bind CD36 analysed across a parasite genome" Mol Microbiol 47: 1265–1278. doi:10.1046/j.1365-2958.2003.03378.x
- ^ Buckee, CO; Recker, M (2012). Antia, Rustom (ed.). "Evolution of the multi-domain structures of virulence genes in the human malaria parasite, Plasmodium falciparum". PLoS Comput Biol. 8 (4): e1002451. doi:10.1371/journal.pcbi.1002451.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: unflagged free DOI (link) - ^ Zilversmit MM, Chase EK, Chen DS, Awadalla P, Day KP, McVean G (2013) Hypervariable antigen genes in malaria have ancient roots. BMC Evol Biol 13(1) 110
- ^ Fastman, Y; Noble, R; Recker, M; Dzikowski, R (2012). Templeton, Thomas J (ed.). "Erasing the epigenetic memory and beginning to switch-The onset of antigenic switching of var genes in Plasmodium falciparum". PLoS ONE. 7 (3): e34168. doi:10.1371/journal.pone.0034168.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Volz JC, Bártfai R, Petter M, Langer C, Josling GA, Tsuboi T, Schwach F, Baum J, Rayner JC, Stunnenberg HG, Duffy MF, Cowman AF (2012) PfSET10, a Plasmodium falciparum methyltransferase, maintains the active var gene in a poised state during parasite division. Cell Host Microbe 11(1) 7-18
- ^ Deshmukh AS, Srivastava S, Herrmann S, Gupta A, Mitra P, Gilberger TW, Dhar SK (2012) The role of N-terminus of Plasmodium falciparum ORC1 in telomeric localization and var gene silencing. Nucleic Acids Res
- ^ Avraham I, Schreier J, Dzikowski R (2012) Insulator-like pairing elements regulate silencing and mutually exclusive expression in the malaria parasite Plasmodium falciparum. Proc Natl Acad Sci USA
- ^ Bancells C, Deitsch KW (2013) A molecular switch in the efficiency of translation reinitiation controls expression of var2csa, a gene implicated in pregnancy associated malaria. Mol Microbiol doi:10.1111/mmi.12379
- ^ Noble R, Christodoulou Z, Kyes S, Pinches R, Newbold CI, Recker M (2013) The antigenic switching network of Plasmodium falciparum and its implications for the immuno-epidemiology of malaria. Elife 2:e01074. doi:10.7554/eLife.01074
- ^ Mayer C, Slater L, Erat MC, Konrat R, Vakonakis I (2012) Structural analysis of the Plasmodium falciparum Erythrocyte membrane protein 1 (PfEMP1) intracellular domain reveals a conserved interactionepitope. J Biol Chem
- ^ Brown A, Turner L, Christoffersen S, Andrews KA, Szestak T, Zhao Y, Larsen S, Craig AG, Higgins MK (2013) Molecular architecture of a complex between an adhesion protein from the malaria parasite and intracellular adhesion molecule 1. J Biol Chem
- ^ Carvalho PA, Diez-Silva M, Chen H, Dao M, Suresh S (2013) Cytoadherence of erythrocytes invaded by Plasmodium falciparum: Quantitative contact-probing of a human malaria receptor. Acta Biomater pii: S1742-7061(13)00034-2. doi:10.1016/j.actbio.2013.01.019
- ^ Angeletti D, Albrecht L, Blomqvist K, Quintana Mdel P, Akhter T, Bächle SM, Sawyer A, Sandalova T, Achour A, Wahlgren M, Moll K (2012) Plasmodium falciparum rosetting epitopes converge in the SD3-loop of PfEMP1-DBL1α" PLoS One 7(12) e50758. doi:10.1371/journal.pone.0050758
- ^ Marsh K, Otoo L, Hayes RJ, Carson DC, Greenwood BM (1989) Antibodies to blood stage antigens of Plasmodium falciparum in rural Gambians and their relation to protection against infection" Trans R Soc Trop Med Hyg 83: 293–303
- ^ Bertin GI, Lavstsen T, Guillonneau F, Doritchamou J, Wang CW, Jespersen JS, Ezimegnon S, Fievet N, Alao MJ, Lalya F, Massougbodji A, Ndam NT, Theander TG, Deloron P (2013) Expression of the Domain Cassette 8 Plasmodium falciparum erythrocyte membrane protein 1 is associated with cerebral malaria in Benin" PLoS One 8(7) e68368. doi:10.1371/journal.pone.0068368
- ^ Turner L, Lavstsen T, Berger SS, Wang CW, Petersen JE, Avril M, Brazier AJ, Freeth J, Jespersen JS, Nielsen MA, Magistrado P, Lusingu J, Smith JD, Higgins MK, Theander TG (2013) Severe malaria is associated with parasite binding to endothelial protein C receptor. Nature doi:10.1038/nature12216
- ^ Rorick MM, Rask TS, Baskerville EB, Day KP, Pascual M. Homology blocks of Plasmodium falciparum var genes and clinically distinct forms of severe malaria in a local population. BMC Microbiol 13(1):244
- ^ Mwakalinga SB, Wang CW, Bengtsson DC, Turner L, Dinko B, Lusingu JP, Arnot DE, Sutherland CJ, Theander TG, Lavstsen T (2012) Expression of a type B RIFIN in Plasmodium falciparum merozoites and gametes. Malar J 11(1) 429
- ^ Sanyal S, Egée S, Bouyer G, Perrot S, Safeukui I, Bischoff E, Buffet P, Deitsch KW, Mercereau-Puijalon O, David PH, Templeton TJ, Lavazec C (2011). "Plasmodium falciparum STEVOR proteins impact erythrocyte mechanical properties". Blood.
{{cite journal}}: CS1 maint: multiple names: authors list (link)
Additional material
- Jewett , Sibley (2003). "Aldolase forms a bridge between cell surface adhesins and the actin cytoskeleton in apicomplexan parasites". Mol Cell. 11 (4): 885–94. doi:10.1016/S1097-2765(03)00113-8. PMID 12718875.
{{cite journal}}:|first2=missing|last2=(help) - Bergman (2003). "Myosin A tail domain interacting protein (MTIP) localizes to the inner membrane complex of Plasmodium sporozoites". J Cell Sci. 116 (1): 39–49. doi:10.1242/jcs.00194.
{{cite journal}}: Invalid|display-authors=1(help) - Baum (2005). "Invasion by P. falciparum merozoites suggests a hierarchy of molecular interactions". PLoS Pathog. 1 (4): e37. doi:10.1371/journal.ppat.0010037.
{{cite journal}}:|first2=missing|last2=(help);|first3=missing|last3=(help);|first4=missing|last4=(help);|first5=missing|last5=(help)CS1 maint: unflagged free DOI (link) - Bosch (2007). "The closed MTIP-myosin A-tail complex from the malaria parasite invasion machinery". J Mol Biol. 372 (1): 77–88. doi:10.1016/j.jmb.2007.06.016. PMC 2702245. PMID 17628590.
{{cite journal}}:|first2=missing|last2=(help);|first3=missing|last3=(help);|first4=missing|last4=(help);|first5=missing|last5=(help);|first6=missing|last6=(help); Invalid|display-authors=1(help) - Bosch (2007). "Aldolase provides an unusual binding site for thrombospondin-related anonymous protein in the invasion machinery of the malaria parasite". Proc Natl Acad Sci USA. 104 (17): 7015–20. doi:10.1073/pnas.0605301104. PMC 1855406. PMID 17426153.
{{cite journal}}:|first2=missing|last2=(help);|first3=missing|last3=(help);|first4=missing|last4=(help);|first5=missing|last5=(help);|first6=missing|last6=(help);|first7=missing|last7=(help);|first8=missing|last8=(help);|first9=missing|last9=(help) - Daher , Soldati-Favre (2009). "Mechanisms controlling glideosome function in apicomplexans". Cur Opin Micro. 12 (4): 408–414. doi:10.1016/j.mib.2009.06.008.
{{cite journal}}:|first2=missing|last2=(help) - Sibley (2010). "How apicomplexan parasites move in and out of cells". Cur Opin Biotech. 21 (5): 592–598. doi:10.1016/j.copbio.2010.05.009.
- Hain and Bosch (2013). "Autophagy in Plasmodium, a multifunctional pathway?". CSBJ. 8 (11). doi:10.5936/csbj.201308002.
{{cite journal}}:|first2=missing|last2=(help) - Boucher and Bosch (2013). "Development of a multifunctional tool for drug screening against plasmodial protein-protein interactions via surface plasmon resonance. |". J. Mol. Recognit. 26 (10): 496–500. doi:10.1002/jmr.2292. PMID 23996492.
{{cite journal}}:|first2=missing|last2=(help)
External links
- MR4, The NIAID funded Malaria Research and Reference Reagent Resource Center
- PlasmoDB
- GeneDB
- Malaria IDC Strain Comparison Database
- Malaria IDC Transcriptome Database
- Malaria Parasite Metabolic Pathways
- ApiCyc
- Library of Apicomplexan metabolic pathways
