ATPase
| Adenosinetriphosphatase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC no. | 3.6.1.3 | ||||||||
| CAS no. | 9000-83-3 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
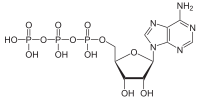
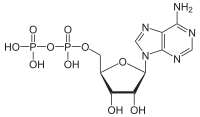
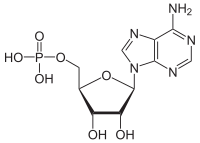
ATPases (EC 3.6.1.3, adenylpyrophosphatase, ATP monophosphatase, triphosphatase, SV40 T-antigen, adenosine 5'-triphosphatase, ATP hydrolase, complex V (mitochondrial electron transport), (Ca2+ + Mg2+)-ATPase, HCO3−-ATPase, adenosine triphosphatase) are a class of enzymes that catalyze the decomposition of ATP into ADP and a free phosphate ion[1][2][3][4][5][6] or the inverse reaction. This dephosphorylation reaction releases energy, which the enzyme (in most cases) harnesses to drive other chemical reactions that would not otherwise occur. This process is widely used in all known forms of life.
Some such enzymes are integral membrane proteins (anchored within biological membranes), and move solutes across the membrane, typically against their concentration gradient. These are called transmembrane ATPases.
Functions

Transmembrane ATPases import many of the metabolites necessary for cell metabolism and export toxins, wastes, and solutes that can hinder cellular processes. An important example is the sodium-potassium exchanger (or Na+/K+ATPase) that maintains the cell membrane potential. And another example is the hydrogen potassium ATPase (H+/K+ATPase or gastric proton pump) that acidifies the contents of the stomach. ATPase is genetically conserved in animals; therefore, cardenolides which are toxic steroids produced by plants that act on ATPases, make general and effective animal toxins that act dose dependently.[7]
Besides exchangers, other categories of transmembrane ATPase include co-transporters and pumps (however, some exchangers are also pumps). Some of these, like the Na+/K+ATPase, cause a net flow of charge, but others do not. These are called "electrogenic" and "nonelectrogenic" transporters, respectively.
Mechanism
The coupling between ATP hydrolysis and transport is more or less a strict chemical reaction, in which a fixed number of solute molecules are transported for each ATP molecule that is hydrolyzed; for example, 3 Na+ ions out of the cell and 2 K+ ions inward per ATP hydrolyzed, for the Na+/K+ exchanger.
Transmembrane ATPases harness the chemical potential energy of ATP, because they perform mechanical work: they transport solutes in a direction opposite to their thermodynamically preferred direction of movement—that is, from the side of the membrane where they are in low concentration to the side where they are in high concentration. This process is considered active transport.
For example, the blocking of the vesicular H+-ATPases would increase the pH inside vesicles and decrease the pH of the cytoplasm.
Transmembrane ATP synthases
The ATP synthase of mitochondria and chloroplasts is an anabolic enzyme that harnesses the energy of a transmembrane proton gradient as an energy source for adding an inorganic phosphate group to a molecule of adenosine diphosphate (ADP) to form a molecule of adenosine triphosphate (ATP).
This enzyme works when a proton moves down the concentration gradient, giving the enzyme a spinning motion. This unique spinning motion bonds ADP and P together to create ATP.
ATP synthase can also function in reverse, that is, use energy released by ATP hydrolysis to pump protons against their electrochemical gradient.
Classification
There are different types of ATPases, which can differ in function (ATP synthesis and/or hydrolysis), structure (F-, V- and A-ATPases contain rotary motors) and in the type of ions they transport.
- Rotary ATPases[8][9]
- F-ATPases (F1FO-ATPases) in mitochondria, chloroplasts and bacterial plasma membranes are the prime producers of ATP, using the proton gradient generated by oxidative phosphorylation (mitochondria) or photosynthesis (chloroplasts).
- V-ATPases (V1VO-ATPases) are primarily found in eukaryotic vacuoles, catalysing ATP hydrolysis to transport solutes and lower pH in organelles like proton pump of lysosome.
- A-ATPases (A1AO-ATPases) are found in Archaea and some extremophilic bacteria. They are arranged like V-ATPases, but function like F-ATPases mainly as ATP synthases.
- P-ATPases (E1E2-ATPases) are found in bacteria, fungi and in eukaryotic plasma membranes and organelles, and function to transport a variety of different ions across membranes.
- E-ATPases are cell-surface enzymes that hydrolyze a range of NTPs, including extracellular ATP.[citation needed]
- AAA proteins are a family of ring-shaped P-loop NTPases.
P-ATPase
P-ATPases (sometime known as E1-E2 ATPases) are found in bacteria and also in eukaryotic plasma membranes and organelles. Its name is due to short time attachment of inorganic phosphate at the aspartate residues at the time of activation. Function of P-ATPase is to transport a variety of different compounds, like ions and phospholipids, across a membrane using ATP hydrolysis for energy. There are many different classes of P-ATPases, which transports a specific type of ion. P-ATPases may be composed of one or two polypeptides, and can usually take two main conformations, E1 and E2.
Human genes
- Na+/K+ transporting: ATP1A1, ATP1A2, ATP1A3, ATP1A4, ATP1B1, ATP1B2, ATP1B3, ATP1B4
- Ca++ transporting: ATP2A1, ATP2A2, ATP2A3, ATP2B1, ATP2B2, ATP2B3, ATP2B4, ATP2C1, ATP2C2
- Mg++ transporting: ATP3
- H+/K+ exchanging: ATP4A
- H+ transporting, mitochondrial: ATP5A1, ATP5B, ATP5C1, ATP5C2, ATP5D, ATP5E, ATP5F1, ATP5G1, ATP5G2, ATP5G3, ATP5H, ATP5I, ATP5J, ATP5J2, ATP5L, ATP5L2, ATP5O, ATP5S
- H+ transporting, lysosomal: ATP6AP1, ATP6AP2, ATP6V1A, ATP6V1B1, ATP6V1B2, ATP6V1C1, ATP6V1C2, ATP6V1D, ATP6V1E1, ATP6V1E2, ATP6V1F, ATP6V1G1, ATP6V1G2, ATP6V1G3, ATP6V1H, ATP6V0A1, ATP6V0A2, ATP6V0A4, ATP6V0B, ATP6V0C, ATP6V0D1, ATP6V0D2, ATP6V0E
- Cu++ transporting: ATP7A, ATP7B
- Class I, type 8: ATP8A1, ATP8B1, ATP8B2, ATP8B3, ATP8B4
- Class II, type 9: ATP9A, ATP9B
- Class V, type 10: ATP10A, ATP10B, ATP10D
- Class VI, type 11: ATP11A, ATP11B, ATP11C
- H+/K+ transporting, nongastric: ATP12A
- type 13: ATP13A1, ATP13A2, ATP13A3, ATP13A4, ATP13A5
See also
References
- ^ Geider K, Hoffmann-Berling H (1981). "Proteins controlling the helical structure of DNA". Annual Review of Biochemistry. 50: 233–60. doi:10.1146/annurev.bi.50.070181.001313. PMID 6267987.
- ^ Kielley WW (1961). "Myosin adenosine triphosphatase". In Boyer PD, Lardy H, Myrbäck K (eds.). The Enzymes. Vol. 5 (2nd ed.). New York: Academic Press. pp. 159–168.
- ^ Martin SS, Senior AE (November 1980). "Membrane adenosine triphosphatase activities in rat pancreas". Biochimica et Biophysica Acta (BBA) - Biomembranes. 602 (2): 401–18. doi:10.1016/0005-2736(80)90320-x. PMID 6252965.
- ^ Njus D, Knoth J, Zallakian M (1981). "Proton-linked transport in chromaffin granules". Current Topics in Bioenergetics. 11: 107–147. doi:10.1016/B978-0-12-152511-8.50010-4.
- ^ Riley MV, Peters MI (June 1981). "The localization of the anion-sensitive ATPase activity in corneal endothelium". Biochimica et Biophysica Acta (BBA) - Biomembranes. 644 (2): 251–6. doi:10.1016/0005-2736(81)90382-5. PMID 6114746.
- ^ Tjian R (1981). "Regulation of viral transcription and DNA replication by the SV40 large T antigen". Current Topics in Microbiology and Immunology. 93: 5–24. doi:10.1007/978-3-642-68123-3_2. ISBN 978-3-642-68125-7. PMID 6269805.
- ^ Dobler S, Dalla S, Wagschal V, Agrawal AA (August 2012). "Community-wide convergent evolution in insect adaptation to toxic cardenolides by substitutions in the Na,K-ATPase". Proceedings of the National Academy of Sciences of the United States of America. 109 (32): 13040–5. doi:10.1073/pnas.1202111109. PMC 3420205. PMID 22826239.
- ^ Stewart AG, Laming EM, Sobti M, Stock D (April 2014). "Rotary ATPases--dynamic molecular machines". Current Opinion in Structural Biology. 25: 40–8. doi:10.1016/j.sbi.2013.11.013. PMID 24878343.
- ^ Kühlbrandt W, Davies KM (January 2016). "Rotary ATPases: A New Twist to an Ancient Machine". Trends in Biochemical Sciences. 41 (1): 106–116. doi:10.1016/j.tibs.2015.10.006. PMID 26671611.
External links
- "ATP synthase - a splendid molecular machine"
- ATPase at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- Electron microscopy structures of ATPases from the EM Data Bank(EMDB)
