Calnexin
Calnexin (CNX) is a 67kDa integral protein (that appears variously as a 90kDa, 80kDa, or 75kDa band on western blotting depending on the source of the antibody) of the endoplasmic reticulum (ER). It consists of a large (50 kDa) N-terminal calcium-binding lumenal domain, a single transmembrane helix and a short (90 residues), acidic cytoplasmic tail.[5]
Function
Calnexin is a chaperone, characterized by assisting protein folding and quality control, ensuring that only properly folded and assembled proteins proceed further along the secretory pathway. It specifically acts to retain unfolded or unassembled N-linked glycoproteins in the ER.[6]
Calnexin binds only those N-glycoproteins that have GlcNAc2Man9Glc1 oligosaccharides.[7] These monoglucosylated oligosaccharides result from the trimming of two glucose residues by the sequential action of two glucosidases, I and II. Glucosidase II can also remove the third and last glucose residue. If the glycoprotein is not properly folded, an enzyme called UGGT (for UDP-glucose:glycoprotein glucosyltransferase) will add the glucose residue back onto the oligosaccharide thus regenerating the glycoprotein's ability to bind to calnexin.[8] The improperly-folded glycoprotein chain thus loiters in the ER and the expression of EDEM/Htm1p [9][10][11] which eventually sentences the underperforming glycoprotein to degradation by removing one of the nine mannose residues. The mannose lectin Yos-9 (OS-9 in humans) marks and sorts misfolded glycoproteins for degradation. Yos-9 recognizes mannose residues exposed after α-mannosidase removal of an outer mannose of misfolded glycoproteins.[12]
Calnexin associates with the protein folding enzyme ERp57[13] to catalyze glycoprotein specific disulfide bond formation and also functions as a chaperone for the folding of MHC class I α-chain in the membrane of the ER. As newly synthesized MHC class I α-chains enter the endoplasmic reticulum, calnexin binds on to them retaining them in a partly folded state.[14]
After the β2-microglobulin binds to the MHC class I peptide-loading complex (PLC), calreticulinand ERp57 take over the job of chaperoning the MHC class I protein while the tapasin links the complex to the transporter associated with antigen processing (TAP) complex. This association prepares the MHC class I for binding an antigen for presentation on the cell surface.
A prolonged association of calnexin with mutant misfolded PMP22 known to cause Charcot-Marie-Tooth Disease [15] leads to the sequestration, degradation and inability of PMP22 to traffic to the Schwann cell surface for myelination. After repeated rounds of calnexin binding, mutant PMP22 is modified by ubiquitin for degradation by the proteasome as well as a Golgi to ER retrieval pathway to return any misfolded PMP22 that escaped from the ER to the Golgi apparatus.[16]
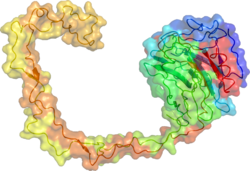
The x-ray crystal structure of calnexin revealed a globular lectin domain and a long hydrophobic arm extending out.[17]
Cofactors
ATP and calcium ions are cofactors involved in substrate binding for calnexin.[18]
References
- ^ a b c ENSG00000127022 GRCh38: Ensembl release 89: ENSG00000283777, ENSG00000127022 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000020368 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ Wada I, Rindress D, Cameron PH, Ou WJ, Doherty JJ 2nd, Louvard D, Bell AW, Dignard D, Thomas DY, Bergeron JJ (1991). "SSR alpha and associated calnexin are major calcium binding proteins of the endoplasmic reticulum membrane". J Biol Chem. 226 (29): 19599–610. PMID 1918067.
- ^ Ou WJ, Cameron PH, Thomas DY, Bergeron JJ (1993). "Association of folding intermediates of glycoproteins". Nature. 364 (644): 771–6. doi:10.1038/364771a0. PMID 8102790.
- ^ Hammond C, Braakman I, Helenius A (1984). "Role of N-linked oligosaccharide recognition, glucose trimming, and calnexin in glycoprotein folding and quality control". Proc Natl Acad Sci USA. 91 (3): 913–7. doi:10.1073/pnas.91.3.913. PMC 521423. PMID 8302866.
- ^ Gañán S, Cazzulo JJ, Parodi AJ (1991). "A major proportion of N-glycoproteins are transiently glucosylated in the endoplasmic reticulum". Biochemistry. 30 (12): 3098–104. doi:10.1021/bi00226a017. PMID 1826090.
- ^ Jacob CA, Bodmer D, Spirig U, Battig P, Marcil A, Dignard D, Bergeron JJ, Thomas DY, Aebi M (2001). "Htm1p, a mannosidase-like protein, is involved in glycoprotein degradation in yeast". EMBO Rep. 2 (5): 423–30. doi:10.1093/embo-reports/kve089. PMC 1083883. PMID 11375935.
- ^ Hosokawa N, Wada I, Hasegawa K, Yorihuzi T, Tremblay LO, Herscovics A, Nagata K (2001). "A novel ER alpha-mannosidase-like protein accelerates ER-associated degradation". EMBO Rep. 2 (5): 415–2. doi:10.1093/embo-reports/kve084. PMC 1083879. PMID 11375934.
- ^ Lee AH, Iwakoshi NN, Glimcher LH (2003). "XBP-1 regulates a subset of endoplasmic reticulum chaperone genes in the unfolded protein response". Mol Cell Biol. 23 (21): 5448–59. doi:10.1128/mcb.23.21.7448-7459.2003. PMC 207643. PMID 14559994.
- ^ Quan EM, Kamiya D, Denic V, Weibezahn J, Kato K, Weissman JS (2008). "Defining the glycan destruction signal for endoplasmic reticulum-associated degradation". Mol Cell. 32 (6): 870–7. doi:10.1016/j.molcel.2008.11.017. PMC 2873636. PMID 19111666.
- ^ Zapun A, Darby NJ, Tessier DC, Michalak M, Bergeron JJ, Thomas DY (1998). "Enhanced catalysis of ribonuclease B folding by the interaction of calnexin or calreticulin with ERp57". J Biol Chem. 273 (211): 6009–12. doi:10.1074/jbc.273.11.6009. PMID 9497314.
- ^ Bergeron JJ, Brenner MB, Thomas DY, Williams DB (1994). "Calnexin: a membrane-bound chaperone of the endoplasmic reticulum". Trends Biochem Sci. 19 (3): 124–8. doi:10.1016/0968-0004(94)90205-4. PMID 8203019.
- ^ Dickson KM, Bergeron JJ, Shames I, Colby J, Nguyen DT, Chevet E, Thomas DY, Snipes GJ (2002). "Association of calnexin with mutant peripheral myelin protein-22 ex vivo: a basis for "gain-of-function" ER diseases". Proc Natl Acad Sci USA. 99 (15): 9852–7. doi:10.1073/pnas.152621799. PMC 125041. PMID 12119418.
- ^ Hara T, Hashimoto Y, Akuzawa T, Hirai R, Kobayashi H, Sato K (2014). "Rer1 and calnexin regulate endoplasmic reticulum retention of a peripheral myelin protein 22 mutant that causes type 1A Charcot-Marie-Tooth disease". Sci Rep. 4: 1–11. doi:10.1038/srep06992. PMC 4227013. PMID 25385046.
- ^ Schrag JD, Bergeron JJ, Li Y, Borisova S, Hahn M, Thomas DY, Cygler M (2001). "The structure of calnexin, an ER chaperone involved in quality control of protein folding". Mol Cell. 8 (3): 633–44. doi:10.1016/s1097-2765(01)00318-5. PMID 11583625.
- ^ Ou WJ, Bergeron JJ, Li Y, Kang CY, Thomas DY (1995). "Conformational changes induced in the endoplasmic reticulum luminal domain of calnexin by Mg-ATP and Ca2+". J Biol Chem. 270 (30): 18051–9. doi:10.1074/jbc.270.30.18051. PMID 7629114.
External links
- Calnexin at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
Further reading
- Benyair R, Ron E, Lederkremer GZ (2011). Protein quality control, retention, and degradation at the endoplasmic reticulum. International Review of Cell and Molecular Biology. Vol. 292. pp. 197–280. doi:10.1016/B978-0-12-386033-0.00005-0. ISBN 9780123860330. PMID 22078962.
- Del Bem LE (Feb 2011). "The evolutionary history of calreticulin and calnexin genes in green plants". Genetica. 139 (2): 225–9. doi:10.1007/s10709-010-9544-y. PMID 21222018.
- Kleizen B, Braakman I (Aug 2004). "Protein folding and quality control in the endoplasmic reticulum". Current Opinion in Cell Biology. 16 (4): 343–9. doi:10.1016/j.ceb.2004.06.012. hdl:1874/5106. PMID 15261665.
- Rasmussen HH, van Damme J, Puype M, Gesser B, Celis JE, Vandekerckhove J (Dec 1992). "Microsequences of 145 proteins recorded in the two-dimensional gel protein database of normal human epidermal keratinocytes". Electrophoresis. 13 (12): 960–9. doi:10.1002/elps.11501301199. PMID 1286667.
- Galvin K, Krishna S, Ponchel F, Frohlich M, Cummings DE, Carlson R, Wands JR, Isselbacher KJ, Pillai S, Ozturk M (Sep 1992). "The major histocompatibility complex class I antigen-binding protein p88 is the product of the calnexin gene". Proceedings of the National Academy of Sciences of the United States of America. 89 (18): 8452–6. doi:10.1073/pnas.89.18.8452. PMC 49938. PMID 1326756.
- Pind S, Riordan JR, Williams DB (Apr 1994). "Participation of the endoplasmic reticulum chaperone calnexin (p88, IP90) in the biogenesis of the cystic fibrosis transmembrane conductance regulator". The Journal of Biological Chemistry. 269 (17): 12784–8. PMID 7513695.
- Honoré B, Rasmussen HH, Celis A, Leffers H, Madsen P, Celis JE (1992). "The molecular chaperones HSP28, GRP78, endoplasmin, and calnexin exhibit strikingly different levels in quiescent keratinocytes as compared to their proliferating normal and transformed counterparts: cDNA cloning and expression of calnexin". Electrophoresis. 15 (3–4): 482–90. doi:10.1002/elps.1150150166. PMID 8055875.
- Tjoelker LW, Seyfried CE, Eddy RL, Byers MG, Shows TB, Calderon J, Schreiber RB, Gray PW (Mar 1994). "Human, mouse, and rat calnexin cDNA cloning: identification of potential calcium binding motifs and gene localization to human chromosome 5". Biochemistry. 33 (11): 3229–36. doi:10.1021/bi00177a013. PMID 8136357.
- Lenter M, Vestweber D (Apr 1994). "The integrin chains beta 1 and alpha 6 associate with the chaperone calnexin prior to integrin assembly". The Journal of Biological Chemistry. 269 (16): 12263–8. PMID 8163531.
- Rajagopalan S, Xu Y, Brenner MB (Jan 1994). "Retention of unassembled components of integral membrane proteins by calnexin". Science. 263 (5145): 387–90. doi:10.1126/science.8278814. PMID 8278814.
- David V, Hochstenbach F, Rajagopalan S, Brenner MB (May 1993). "Interaction with newly synthesized and retained proteins in the endoplasmic reticulum suggests a chaperone function for human integral membrane protein IP90 (calnexin)". The Journal of Biological Chemistry. 268 (13): 9585–92. PMID 8486646.
- Bellovino D, Morimoto T, Tosetti F, Gaetani S (Jan 1996). "Retinol binding protein and transthyretin are secreted as a complex formed in the endoplasmic reticulum in HepG2 human hepatocarcinoma cells". Experimental Cell Research. 222 (1): 77–83. doi:10.1006/excr.1996.0010. PMID 8549676.
- Otteken A, Moss B (Jan 1996). "Calreticulin interacts with newly synthesized human immunodeficiency virus type 1 envelope glycoprotein, suggesting a chaperone function similar to that of calnexin". The Journal of Biological Chemistry. 271 (1): 97–103. doi:10.1074/jbc.271.1.97. PMID 8550632.
- Devergne O, Hummel M, Koeppen H, Le Beau MM, Nathanson EC, Kieff E, Birkenbach M (Feb 1996). "A novel interleukin-12 p40-related protein induced by latent Epstein-Barr virus infection in B lymphocytes". Journal of Virology. 70 (2): 1143–53. PMC 189923. PMID 8551575.
- Andersson B, Wentland MA, Ricafrente JY, Liu W, Gibbs RA (Apr 1996). "A "double adaptor" method for improved shotgun library construction". Analytical Biochemistry. 236 (1): 107–13. doi:10.1006/abio.1996.0138. PMID 8619474.
- van Leeuwen JE, Kearse KP (Apr 1996). "Calnexin associates exclusively with individual CD3 delta and T cell antigen receptor (TCR) alpha proteins containing incompletely trimmed glycans that are not assembled into multisubunit TCR complexes". The Journal of Biological Chemistry. 271 (16): 9660–5. doi:10.1074/jbc.271.16.9660. PMID 8621641.
- Oliver JD, Hresko RC, Mueckler M, High S (Jun 1996). "The glut 1 glucose transporter interacts with calnexin and calreticulin". The Journal of Biological Chemistry. 271 (23): 13691–6. doi:10.1074/jbc.271.23.13691. PMID 8662691.
- Li Y, Bergeron JJ, Luo L, Ou WJ, Thomas DY, Kang CY (Sep 1996). "Effects of inefficient cleavage of the signal sequence of HIV-1 gp 120 on its association with calnexin, folding, and intracellular transport". Proceedings of the National Academy of Sciences of the United States of America. 93 (18): 9606–11. doi:10.1073/pnas.93.18.9606. PMC 38475. PMID 8790377.
- Trombetta ES, Simons JF, Helenius A (Nov 1996). "Endoplasmic reticulum glucosidase II is composed of a catalytic subunit, conserved from yeast to mammals, and a tightly bound noncatalytic HDEL-containing subunit". The Journal of Biological Chemistry. 271 (44): 27509–16. doi:10.1074/jbc.271.44.27509. PMID 8910335.
- Tatu U, Helenius A (Feb 1997). "Interactions between newly synthesized glycoproteins, calnexin and a network of resident chaperones in the endoplasmic reticulum". The Journal of Cell Biology. 136 (3): 555–65. doi:10.1083/jcb.136.3.555. PMC 2134297. PMID 9024687.
- Wiest DL, Bhandoola A, Punt J, Kreibich G, McKean D, Singer A (Mar 1997). "Incomplete endoplasmic reticulum (ER) retention in immature thymocytes as revealed by surface expression of "ER-resident" molecular chaperones". Proceedings of the National Academy of Sciences of the United States of America. 94 (5): 1884–9. doi:10.1073/pnas.94.5.1884. PMC 20012. PMID 9050874.
- Yu W, Andersson B, Worley KC, Muzny DM, Ding Y, Liu W, Ricafrente JY, Wentland MA, Lennon G, Gibbs RA (Apr 1997). "Large-scale concatenation cDNA sequencing". Genome Research. 7 (4): 353–8. doi:10.1101/gr.7.4.353. PMC 139146. PMID 9110174.