Catenin beta-1: Difference between revisions
mNo edit summary |
→The beta-catenin destruction complex: typo corrected |
||
| Line 37: | Line 37: | ||
[[File:Beta-catenin-destruction-complex.png|thumb|left|upright=1.5|Simplified structure of the beta-catenin destruction complex. Note the high proportion of intrinsically disordered segments in the axin and APC proteins.]] |
[[File:Beta-catenin-destruction-complex.png|thumb|left|upright=1.5|Simplified structure of the beta-catenin destruction complex. Note the high proportion of intrinsically disordered segments in the axin and APC proteins.]] |
||
But even ''axin'' does not act alone. Through its N-terminal [[Regulator of G protein signalling|regulator of G-protein signaling]] (RGS) domain, it recruits the [[adenomatous polyposis coli]] (APC) protein. ''APC'' is like a huge "Christmas tree": with a multitude of β-catenin binding motifs (one ''APC'' molecule alone possesses |
But even ''axin'' does not act alone. Through its N-terminal [[Regulator of G protein signalling|regulator of G-protein signaling]] (RGS) domain, it recruits the [[adenomatous polyposis coli]] (APC) protein. ''APC'' is like a huge "Christmas tree": with a multitude of β-catenin binding motifs (one ''APC'' molecule alone possesses 11 such motifs <ref>{{cite journal|last=Minde|first=David P|coauthors=Anvarian, Zeinab; Rüdiger, Stefan GD; Maurice, Madelon M|title=Messing up disorder: how do missense mutations in the tumor suppressor protein APC lead to cancer?|journal=Molecular Cancer|date=1 January 2011|volume=10|issue=1|pages=101|doi=10.1186/1476-4598-10-101|pmid=21859464|pmc=3170638}}</ref>), it may collect as many β-catenin molecules as possible.<ref name="pmid16753179">{{cite journal |author=Liu J, Xing Y, Hinds TR, Zheng J, Xu W |title=The third 20 amino acid repeat is the tightest binding site of APC for beta-catenin |journal=J. Mol. Biol. |volume=360 |issue=1 |pages=133–44 |date=June 2006 |pmid=16753179 |doi=10.1016/j.jmb.2006.04.064 |url=}}</ref> ''APC'' can interact with multiple ''axin'' molecules at the same time as it has not one but ''three'' so-called ''SAMP motifs'' (Ser-Ala-Met-Pro) to bind the [[Regulator of G protein signalling|RGS domains]] found in ''axin''. In addition, axin also has the potential to oligomerize through its C-terminal DIX domain. The result is a huge, multimeric protein assembly dedicated to β-catenin phosphorylation. This complex is usually called the ''beta-catenin destruction complex'', although it is distinct from the [[proteosome]] machinery actually responsible for β-catenin degradation.<ref name="pmid17143292">{{cite journal |author=Kimelman D, Xu W |title=beta-catenin destruction complex: insights and questions from a structural perspective |journal=Oncogene |volume=25 |issue=57 |pages=7482–91 |date=December 2006 |pmid=17143292 |doi=10.1038/sj.onc.1210055 |url=}}</ref> It only marks β-catenin molecules for subsequent destruction. |
||
== Wnt signaling and the regulation of destruction == |
== Wnt signaling and the regulation of destruction == |
||
Revision as of 08:30, 15 May 2014
Catenin (cadherin-associated protein), beta 1, 88kDa (the HUGO-approved official symbol, CTNNB1; HGNC ID, HGNC:2514), also called beta-catenin (or β-catenin), is a dual function protein, regulating the coordination of cell–cell adhesion and gene transcription. In humans, the CTNNB1 protein is encoded by the CTNNB1 gene.[1][2] In Drosophila, the homologous protein is called armadillo. β-catenin is a subunit of the cadherin protein complex and acts as an intracellular signal transducer in the Wnt signaling pathway.[3][4][5]
Mutations and overexpression of β-catenin are associated with many cancers, including hepatocellular carcinoma, colorectal carcinoma, lung cancer, malignant breast tumors, ovarian and endometrial cancer.[6] β-catenin is regulated and destroyed by the beta-catenin destruction complex, and in particular by the adenomatous polyposis coli (APC) protein, encoded by the tumour-suppressing APC gene. Therefore genetic mutation of the APC gene is also strongly linked to cancers, and in particular colorectal cancer resulting from familial adenomatous polyposis (FAP).
Discovery
Beta-catenin was initially discovered in the early 1990s as a component of a mammalian cell adhesion complex: a protein responsible for cytoplasmatic anchoring of cadherins.[7] But very soon, it was realized that the Drosophila protein armadillo - implicated in mediating the morphogenic effects of Wingless/Wnt - is homologous to the mammalian β-catenin, not just in structure but also in function.[8] Thus beta-catenin became one of the very first examples of moonlighting: a protein performing more than one radically different cellular function.
Protein structure
The core of beta-catenin consists of several, very characteristic repeats of approximately 40 amino acids long. Termed armadillo repeats, all these elements fold together into a single, rigid protein domain with an elongated shape - called armadillo (ARM) domain. An average armadillo repeat is composed of three alpha helices. The first repeat of β-catenin (near the N-terminus) is slightly different from the others - as it has an elongated helix with a kink, formed by the fusion of helices 1 and 2.[9] Due to the complex shape of individual repeats, the whole ARM domain is not a straight rod: it possesses a slight curvature, so that an outer (convex) and an inner (concave) surface is formed. This inner surface serves as a ligand-binding site for the various interaction partners of the ARM domains.

The segments N-terminal and far C-terminal to the ARM domain do not adopt any structure in solution by themselves. Yet these intrinsically disordered regions play a crucial role in beta-catenin function. The N-terminal disordered region contains a conserved short linear motif responsible for binding of TrCP1 (also known as β-TrCP) E3 ubiquitin ligase - but only when it is phosphorylated. Degradation of β-catenin is thus mediated by this N-terminal segment. The C-terminal region, on the other hand, is a strong transactivator when recruited onto DNA. This segment is not fully disordered: part of the C-terminal extension forms a stable helix that packs against the ARM domain, but may also engage separate binding partners.[10] This small structural element (HelixC) caps the C-terminal end of the ARM domain, shielding its hydrophobic residues. HelixC is not necessary for beta-catenin to function in cell-cell adhesion. On the other hand, it is required for Wnt signaling: possibly to recruit various coactivators. Yet its exact partners among the general transcription complexes are still unknown. Notably, the C-terminal segment of β-catenin can mimic the effects of the entire Wnt pathway if artificially fused to the DNA binding domain of LEF1 transcription factor.[11]
Plakoglobin (also called gamma-catenin) has a strikingly similar architecture to that of beta-catenin. Not only their ARM domains resemble each other in both architecture and ligand binding capacity, but the N-terminal β-TrCP-binding motif is also conserved in plakoglobin, implying common ancestry and shared regulation with β-catenin.[12] However, plakoglobin is a very weak transactivator when bound to DNA - this is probably caused by the divergence of their C-terminal sequences (plakoglobin appears to lack the transactivator motifs, and thus inhibits the Wnt pathway target genes instead of activating them).[13]
Partners binding to the armadillo domain
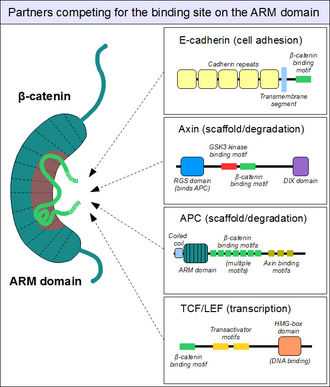
As sketched above, the ARM domain of beta-catenin acts as a platform to which specific linear motifs may bind. Located in structurally diverse partners, the β-catenin binding motifs are typically disordered on their own, and only adopt a rigid structure upon ARM domain engagement - as typical for short linear motifs. However, β-catenin interacting motifs also have a number of peculiar characteristics. First, they might reach or even surpass the length of 30 amino acids in length, and contact the ARM domain on an excessively large surface area. Another unusual feature of these motifs is their frequently high degree of phosphorylation. Such Ser/Thr phosphorylation events greatly enhance the binding of many β-catenin associating motifs to the ARM domain.[14]
Among dedicated partners of beta-catenin we can find the familiar E-cadherin, whose cytoplasmatic tail contacts the ARM domain in a canonical fashion.[15] The scaffold protein axin (two closely related paralogs, axin 1 and axin 2) contains a similar interaction motif on its long, disordered middle segment.[16] Although one molecule of axin only contains a single β-catenin recruitment motif, its partner the Adenomatous Polyposis Coli (APC) protein contains at least 10 (!) such motifs in tandem arrangement, thus capable to interact with several β-catenin molecules at once. Finally, the TCF/LEF transcription factors also feature a β-catenin recruiting motif on their N-terminal disordered segment. Since the surface of the ARM domain can accommodate only one peptide motif at any given time, all these proteins compete for the same cellular pool of β-catenin molecules. This competition is the key to understand how the Wnt signaling pathway works.
However, this "main" binding site on the ARM domain β-catenin is by no means the only one. The first helices of the ARM domain form an additional, special protein-protein interaction pocket: This can accommodate a helix-forming linear motif found in the coactivator BCL9 (or the closely related BCL9L) - an important protein involved in Wnt signaling.[17] Although the precise details are much less clear, it appears that the same site is used by alpha-catenin when beta-catenin is localized to the adherens junctions.[18] Because this pocket is distinct from the ARM domain's "main" binding site, there is no competition between alpha-catenin and E-cadherin or between TCF1 and BCL9, respectively.[19] On the other hand, BCL9 and BCL9L must compete with α-catenin to access β-catenin molecules.[20]
Regulation of degradation through phosphorylation
The cellular level of beta-catenin is mostly controlled by its ubiquitinylation and proteosomal degradation. The E3 ubiquitin ligase TrCP1 (also known as β-TrCP) can recognize β-catenin as its substrate through a short linear motif on the disordered N-terminus. However, this motif (Asp-Ser-Gly-Ile-His-Ser) of β-catenin needs to be phosphorylated on the two serines in order to be capable to bind β-TrCP. Phosphorylation of the motif is performed by Glycogen Synthase Kinase 3 alpha and beta (GSK3α and GSK3β). GSK3s are constitutively active enzymes implicated in several important regulatory processes. There is one requirement, though: substrates of GSK3 need to be pre-phosphorylated four amino acids downstream (C-terminally) of the actual target site. Thus it also requires a "priming kinase" for its activities. In the case of beta-catenin, the most important priming kinase is Casein Kinase I (CKI). Once a serin-threonine rich substrate has been "primed", GSK3 can "walk" across it from C-terminal to N-terminal direction, phosphorylating every 4th serine or threonine residues in a row. This process will result in dual phosphorylation of the aforementioned β-TrCP recognition motif as well.
The beta-catenin destruction complex
For GSK3 to be a highly effective kinase on a substrate, pre-phosphorylation is not enough. There is one additional requirement: Similar to the mitogen-activated protein kinases (MAPKs), substrates need to associate with this enzyme through high-affinity docking motifs. Beta-catenin contains no such motifs, but a special protein: axin does. What is more, its GSK3 docking motif is directly adjacent to a β-catenin binding motif.[16] This way, axin acts as a true scaffold protein, bringing an enzyme (GSK3) together with its substrate (β-catenin) into close physical proximity.

But even axin does not act alone. Through its N-terminal regulator of G-protein signaling (RGS) domain, it recruits the adenomatous polyposis coli (APC) protein. APC is like a huge "Christmas tree": with a multitude of β-catenin binding motifs (one APC molecule alone possesses 11 such motifs [21]), it may collect as many β-catenin molecules as possible.[22] APC can interact with multiple axin molecules at the same time as it has not one but three so-called SAMP motifs (Ser-Ala-Met-Pro) to bind the RGS domains found in axin. In addition, axin also has the potential to oligomerize through its C-terminal DIX domain. The result is a huge, multimeric protein assembly dedicated to β-catenin phosphorylation. This complex is usually called the beta-catenin destruction complex, although it is distinct from the proteosome machinery actually responsible for β-catenin degradation.[23] It only marks β-catenin molecules for subsequent destruction.
Wnt signaling and the regulation of destruction
In resting cells, axin molecules oligomerize with each other through their C-terminal DIX domains, which have two binding interfaces. Thus they can build linear oligomers or even polymers inside the cytoplasm of cells. DIX domains are very unique: the only other protein known to have a DIX domain is Dishevelled. (The single Dsh protein of Drosophila corresponds to three paralogous genes, Dvl1, Dvl2 and Dvl3 in mammals.) Dsh associates with the cytoplasmatic regions of Frizzled receptors with its PDZ and DEP domains. When a Wnt molecule binds to Frizzled, it induces a poorly known cascade of events, that result in the exposure of dishevelled's DIX domain and the creation of a perfect binding site for axin. Axin is then titrated away from its oligomeric assemblies - the β-catenin destruction complex - by Dsh.[24] Once bound to the receptor complex, axin will be rendered incompetent for β-catenin binding and GSK3 activity. Importantly, the cytoplasmatic segments of the Frizzled-associated LRP5 and LRP6 proteins contain GSK3 pseudo-substrate sequences (Pro-Pro-Pro-Ser-Pro-x-Ser), appropriately "primed" (pre-phosphorylated) by CKI, as if it were a true substrate of GSK3. These false target sites greatly inhibit GSK3 activity in a comptetitive manner.[25] This way receptor-bound axin will abolish mediating the phosphorylation of β-catenin. Since beta-catenin is no longer marked for destruction, but continues to be produced, its concentration will increase. Once β-catenin levels rise high enough to saturate all binding sites in the cytoplasm, it will also translocate into the nucleus. Upon engaging the transcription factors LEF1, TCF1, TCF2 or TCF3, β-catenin forces them to disengage their previous partners: Groucho proteins. Unlike Groucho, that recruit transcriptional repressors (e.g. histone-lysine methyltransferases), beta-catenin will bind transcriptional activators, switching on target genes.
Role in cell-cell adhesion

Cell–cell adhesion complexes are essential for the formation of complex animal tissues. β-catenin is part of a protein complex that form the so-called adherens junctions.[26] These cell-cell adhesion complexes are necessary for the creation and maintenance of epithelial cell layers and barriers. As a component of the complex, β-catenin can regulate cell growth and adhesion between cells. It may also be responsible for transmitting the contact inhibition signal that causes cells to stop dividing once the epithelial sheet is complete.[27] The E-cadherin – β-catenin – α-catenin complex is weakly associated to actin filaments. Adherent junctions thus form a dynamic, rather than a stable link to the actin cytoskeleton.[26]
The heart of the adherent junctions are the cadherin proteins. Cadherins form the cell-cell junctional structures known as adherens junctions as well as the desmosomes. Cadherins are capable of homophilic interactions through their extracellular cadherin repeat domains, in a Ca2+-dependent manner: this can hold adjacent epithelial cells together. While in the adherens junction, cadherins recruit β-catenin molecules onto their intracellular regions. β-catenin, in turn, associates with another important protein, α-catenin that directly binds to the actin filaments.[28] This is possible because α-catenin and cadherins bind at distinct sites to β-catenin. The β-catenin - α-catenin complex can thus physically bridge cadherins with the actin cytoskeleton.[29] Organization of the cadherin–catenin complex is additionally regulated through phosphorylation and endocytosis of its components.
Roles in development
Beta-catenin has a central role in directing several developmental processes, as it can directly bind transcription factors and be regulated by a diffusible extracellular substance: Wnt. It acts upon early embryos to induce entire body regions, as well as individual cells in later stages of development. It also regulates physiological regeneration processes.
Early embryonic patterning
Wnt signaling and beta-catenin dependent gene expression plays a critical role during the formation of different body regions in the early embryo. Experimentally modified embryos that do not express this protein will fail to develop mesoderm and initiate gastrulation.[30] During the blastula and gastrula stages, Wnt as well as BMP and FGF pathways will induce the antero-posterior axis formation, regulate the precise placement of the primitive streak (gastrulation and mesoderm formation) as well as the process of neurulation (central nervous system development).[31]
In Xenopus oocytes, β-catenin is initially equally localized to all regions of the egg, but it is targeted for ubiquitination and degradation by the β-catenin destruction complex. Fertilization of the egg causes a rotation of the outer cortical layers, moving clusters of the Frizzled and Dsh proteins closer to the equatorial region. β-catenin will be enriched locally under the influence of Wnt signaling pathway in the cells that inherit this portion of the cytoplasm. It will eventually translocate to the nucleus to bind TCF3 in order to activate several genes that induce dorsal cell characteristics.[32] This signaling results in a region of cells known as the grey crescent, which is a classical organizer of embryonic development. If this region is surgically removed from the embryo, gastrulation does not occur at all. β-Catenin also plays a crucial role in the induction of the blastopore lip, which in turn initiates gastrulation.[33] Inhibition of GSK-3 translation by injection of antisense mRNA may cause a second blastopore and a superfluous body axis to form. A similar effect can result from the overexpression of β-catenin.[34]
Asymmetric cell division
Beta-catenin has also been implicated in regulation of cell fates through asymmetric cell division in the model organism C. elegans. Similarly to the Xenopus oocytes, this is essentially the result of non-equal distribution of Dsh, Frizzled, axin and APC in the cytoplasm of the mother cell.[35]
Stem cell renewal
One of the most important results of Wnt signaling and the elevated level of beta-catenin in certain cell types is the maintenance of pluripotency.[31] In other cell types and developmental stages, β-catenin may promote differentiation, especially towards mesodermal cell lineages.
Epithelial-to-mesenchymal transition
Beta-catenin also acts as a morphogen in later stages of embryonic development. Together with TGF-β, an important role of β-catenin is to induce a morphogenic change in epithelial cells. It induces them to abandon their tight adhesion and assume a more mobile and loosely associated mesenchymal phenotype. During this process, epithelial cells lose expression of proteins like E-cadherin, Zonula occludens 1 (ZO1), and cytokeratin. At the same time they turn on the expression of vimentin, alpha smooth muscle actin (ACTA2), and fibroblast-specific protein 1 (FSP1). They also produce extracellular matrix components, such as type I collagen and fibronectin. Aberrant activation of the Wnt pathway has been implicated in pathological processes such as fibrosis and cancer.[36]
Involvement in cancer

Beta-catenin is a proto-oncogene. Mutations of this gene are commonly found in a variety of cancers: in primary hepatocellular carcinoma, colorectal cancer, ovarial carcinoma, breast cancer, lung cancer and glioblastoma. It has been estimated that approximately 10% of all tissue samples sequenced from all cancers display mutations in the CTNNB1 gene.[37] Most of these mutations cluster on a tiny area of the N-terminal segment of β-catenin: the β-TrCP binding motif. Loss-of-function mutations of this motif essentially make ubiquitinylation and degradation of β-catenin impossible. It will cause β-catenin to translocate to the nucleus without any external stimulus and continuously drive transcription of its target genes. Increased nuclear β-catenin levels have also been noted in basal cell carcinoma (BCC),[38] head and neck squamous cell carcinoma (HNSCC), prostate cancer (CaP),[39] pilomatrixoma (PTR)[40] and medulloblastoma (MDB)[41] These observations may or may not implicate a mutation in the β-catenin gene: other Wnt pathway components can also be faulty.
Similar mutations are also frequently seen in the β-catenin recruiting motifs of APC. Hereditary loss-of-function mutations of APC cause a condition known as Familial Adenomatous Polyposis. Affected individuals develop hundreds of polyps in their large intestine. Most of these polyps are benign in nature, but they have the potential to transform into deadly cancer as time progresses. Somatic mutations of APC in colorectal cancer are also not uncommon.[42] Beta-catenin and APC are among the key genes (together with others, like K-Ras and SMAD4) involved in colorectal cancer development. The potential of β-catenin to change the previously epithelial phenotype of affected cells into an invasive, mesenchyme-like type contributes greatly to metastasis formation.
As a therapeutic target
Due to its involvement in cancer development, inhibition of beta-catenin continues to receive significant attention. But the targeting of the binding site on its armadillo domain is not the simplest task, due to its extensive and relatively flat surface. However, for an efficient inhibition, binding to smaller "hotspots" of this surface is sufficient. This way, a "stapled" helical peptide derived from the natural β-catenin binding motif found in LEF1 was sufficient for the complete inhibition of β-catenin dependent transcription. Recently, several small-molecule compounds have also been developed to target the same, highly positively charged area of the ARM domain (CGP049090, PKF118-310, PKF115-584 and ZTM000990). In addition, β-catenin levels can also be influenced by targeting upstream components of the Wnt pathway as well as the β-catenin destruction complex.[43] The additional N-terminal binding pocket is also important for Wnt target gene activation (required for BCL9 recruitment). This site of the ARM domain can be pharmacologically targeted by carnosic acid, for example.[44] That "auxiliary" site is another attractive target for drug development.[45] Despite intensive preclinical research, no β-catenin inhibitors are available as therapeutic agents yet.
Binding partners of beta-catenin (list)
Beta-catenin has been shown to interact with:
- APC,[46][47][48][49][50][51][52][53]
- AXIN1,[54][55]
- Androgen receptor,[56][57][58][59][60][61]
- CBY1,[62]
- CDH1,[15][47][63][64][65][66][67][68][69][70][71][72][73][74][75][76][77][78][79][80][81][82][83]
- CDH2,[84][85]
- CDH3,[82][86]
- CDK5R1,[87]
- CHUK,[88]
- CTNND1,[47][68]
- CTNNA1,[64][73][89][90][91]
- EGFR,[68][77][92]
- FHL2,[93]
- GSK3B,[49][94]
- HER2/neu,[69][92][95]
- HNF4A,[60]
- IKK2,[88]
- LEF1,[96][97][98][99]
- MAGI1,[78]
- MUC1,[70][100][101][102][103][104][105]
- NR5A1,[106][107]
- PCAF,[108]
- PHF17,[109]
- Plakoglobin,[47][68]
- PTPN14,[110]
- PTPRF,[69][111]
- PTPRK (PTPkappa),[112]
- PTPRT (PTPrho),[113]
- PTPRU (PCP-2),[114][115][116]
- PSEN1,[117][118][119]
- PTK7[120]
- RuvB-like 1,[121]
- SMAD7,[96]
- SMARCA4[122]
- SLC9A3R1,[72]
- USP9X,[123] and
- VE-cadherin.[124][125]
See also
References
- ^ Kraus C, Liehr T, Hülsken J, Behrens J, Birchmeier W, Grzeschik KH, Ballhausen WG (September 1994). "Localization of the human CTNNB1 gene to 3p21: a region implicated in tumor development". Genomics. 23 (1): 272–4. doi:10.1006/geno.1994.1493. PMID 7829088.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ MacDonald BT, Tamai K, He X (July 2009). "Wnt/beta-catenin signaling: components, mechanisms, and diseases". Developmental Cell. 17 (1): 9–26. doi:10.1016/j.devcel.2009.06.016. PMC 2861485. PMID 19619488.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Peifer M, Rauskolb C, Williams M, Riggleman B, Wieschaus E (Apr 1991). "The segment polarity gene armadillo interacts with the wingless signaling pathway in both embryonic and adult pattern formation". Development. 111 (4): 1029–1043. PMID 1879348.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Noordermeer J, Klingensmith, J, Perrimon, N, Nusse, R (Jan 1994). "dishevelled and armadillo act in the wingless signalling pathway in Drosophila". Nature. 367 (6458): 80–83. doi:10.1038/367080a0. PMID 7906389.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Peifer M, Berg S, Reynolds AB (March 1994). "A repeating amino acid motif shared by proteins with diverse cellular roles". Cell. 76 (5): 789–91. doi:10.1016/0092-8674(94)90353-0. PMID 7907279.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ "beta-catenin signaling and cancer". BioEssays. 21 (12): 1021–30. Dec 1999. doi:10.1002/(SICI)1521-1878(199912)22:11021::AID-BIES63.0.CO;2-P. PMID 10580987.
- ^ McCrea PD, Turck CW, Gumbiner B (November 1991). "A homolog of the armadillo protein in Drosophila (plakoglobin) associated with E-cadherin". Science. 254 (5036): 1359–61. doi:10.1126/science.1962194. PMID 1962194.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Kemler R (September 1993). "From cadherins to catenins: cytoplasmic protein interactions and regulation of cell adhesion". Trends Genet. 9 (9): 317–21. PMID 8236461.
- ^ Gottardi CJ, Peifer M (March 2008). "Terminal regions of beta-catenin come into view". Structure. 16 (3): 336–8. doi:10.1016/j.str.2008.02.005. PMC 2329800. PMID 18334207.
- ^ Xing Y, Takemaru K, Liu J, Berndt JD, Zheng JJ, Moon RT, Xu W (March 2008). "Crystal structure of a full-length beta-catenin". Structure. 16 (3): 478–87. doi:10.1016/j.str.2007.12.021. PMID 18334222.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Vleminckx K, Kemler R, Hecht A (March 1999). "The C-terminal transactivation domain of beta-catenin is necessary and sufficient for signaling by the LEF-1/beta-catenin complex in Xenopus laevis". Mech. Dev. 81 (1–2): 65–74. PMID 10330485.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Sadot E, Simcha I, Iwai K, Ciechanover A, Geiger B, Ben-Ze'ev A (April 2000). "Differential interaction of plakoglobin and beta-catenin with the ubiquitin-proteasome system". Oncogene. 19 (16): 1992–2001. doi:10.1038/sj.onc.1203519. PMID 10803460.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Aktary Z, Pasdar M (2012). "Plakoglobin: role in tumorigenesis and metastasis". Int J Cell Biol. 2012: 189521. doi:10.1155/2012/189521. PMC 3312339. PMID 22481945.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Xu W, Kimelman D (October 2007). "Mechanistic insights from structural studies of beta-catenin and its binding partners". J. Cell. Sci. 120 (Pt 19): 3337–44. doi:10.1242/jcs.013771. PMID 17881495.
- ^ a b Huber AH, Weis WI (May 2001). "The structure of the beta-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by beta-catenin". Cell. 105 (3): 391–402. doi:10.1016/S0092-8674(01)00330-0. PMID 11348595. Cite error: The named reference "pmid11348595" was defined multiple times with different content (see the help page).
- ^ a b Xing Y, Clements WK, Kimelman D, Xu W (November 2003). "Crystal structure of a beta-catenin/axin complex suggests a mechanism for the beta-catenin destruction complex". Genes Dev. 17 (22): 2753–64. doi:10.1101/gad.1142603. PMC 280624. PMID 14600025.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Kramps T, Peter O, Brunner E; et al. (April 2002). "Wnt/wingless signaling requires BCL9/legless-mediated recruitment of pygopus to the nuclear beta-catenin-TCF complex". Cell. 109 (1): 47–60. doi:10.1016/S0092-8674(02)00679-7. PMID 11955446.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Pokutta S, Weis WI (March 2000). "Structure of the dimerization and beta-catenin-binding region of alpha-catenin". Mol. Cell. 5 (3): 533–43. doi:10.1016/S1097-2765(00)80447-5. PMID 10882138.
- ^ Sampietro J, Dahlberg CL, Cho US, Hinds TR, Kimelman D, Xu W (October 2006). "Crystal structure of a beta-catenin/BCL9/Tcf4 complex". Mol. Cell. 24 (2): 293–300. doi:10.1016/j.molcel.2006.09.001. PMID 17052462.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Brembeck FH, Schwarz-Romond T, Bakkers J, Wilhelm S, Hammerschmidt M, Birchmeier W (September 2004). "Essential role of BCL9-2 in the switch between beta-catenin's adhesive and transcriptional functions". Genes Dev. 18 (18): 2225–30. doi:10.1101/gad.317604. PMC 517514. PMID 15371335.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Minde, David P (1 January 2011). "Messing up disorder: how do missense mutations in the tumor suppressor protein APC lead to cancer?". Molecular Cancer. 10 (1): 101. doi:10.1186/1476-4598-10-101. PMC 3170638. PMID 21859464.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help)CS1 maint: unflagged free DOI (link) - ^ Liu J, Xing Y, Hinds TR, Zheng J, Xu W (June 2006). "The third 20 amino acid repeat is the tightest binding site of APC for beta-catenin". J. Mol. Biol. 360 (1): 133–44. doi:10.1016/j.jmb.2006.04.064. PMID 16753179.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Kimelman D, Xu W (December 2006). "beta-catenin destruction complex: insights and questions from a structural perspective". Oncogene. 25 (57): 7482–91. doi:10.1038/sj.onc.1210055. PMID 17143292.
- ^ Fiedler M, Mendoza-Topaz C, Rutherford TJ, Mieszczanek J, Bienz M (February 2011). "Dishevelled interacts with the DIX domain polymerization interface of Axin to interfere with its function in down-regulating β-catenin". Proceedings of the National Academy of Sciences of the United States of America. 108 (5): 1937–42. doi:10.1073/pnas.1017063108. PMC 3033301. PMID 21245303.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Metcalfe C, Bienz M (November 2011). "Inhibition of GSK3 by Wnt signalling--two contrasting models". J. Cell. Sci. 124 (Pt 21): 3537–44. doi:10.1242/jcs.091991. PMID 22083140.
- ^ a b Brembeck FH, Rosário M, Birchmeier W (February 2006). "Balancing cell adhesion and Wnt signalin], the key role of β-catenin". Current Opinion in Genetics & Development. 16 (1): 51–9. doi:10.1016/j.gde.2005.12.007. PMID 16377174.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ "Entrez Gene: catenin (cadherin-associated protein)".
- ^ Nelson WJ (April 2008). "Regulation of cell-cell adhesion by the cadherin-catenin complex". Biochem. Soc. Trans. 36 (Pt 2): 149–55. doi:10.1042/BST0360149. PMC 3368607. PMID 18363555.
- ^ Bienz M (January 2005). "β-Catenin: a pivot between cell adhesion and Wnt signalling". Curr. Biol. 15 (2): R64–7. doi:10.1016/j.cub.2004.12.058. PMID 15668160.
- ^ Haegel H, Larue L, Ohsugi M, Fedorov L, Herrenknecht K, Kemler R (November 1995). "Lack of beta-catenin affects mouse development at gastrulation". Development. 121 (11): 3529–37. PMID 8582267.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b Sokol SY (October 2011). "Maintaining embryonic stem cell pluripotency with Wnt signaling". Development. 138 (20): 4341–50. doi:10.1242/dev.066209. PMC 3177306. PMID 21903672.
- ^ Schneider S, Steinbeisser H, Warga RM, Hausen P (July 1996). "Beta-catenin translocation into nuclei demarcates the dorsalizing centers in frog and fish embryos". Mech. Dev. 57 (2): 191–8. PMID 8843396.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Larabell CA, Torres M, Rowning BA, Yost C, Miller JR, Wu M, Kimelman D, Moon RT (March 1997). "Establishment of the dorso-ventral axis in Xenopus embryos is presaged by early asymmetries in beta-catenin that are modulated by the Wnt signaling pathway". J. Cell Biol. 136 (5): 1123–36. PMC 2132470. PMID 9060476.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Kelly GM, Erezyilmaz DF, Moon RT (October 1995). "Induction of a secondary embryonic axis in zebrafish occurs following the overexpression of beta-catenin". Mech. Dev. 53 (2): 261–73. PMID 8562427.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Sawa H (2012). "Control of cell polarity and asymmetric division in C. elegans". Curr. Top. Dev. Biol. 101: 55–76. doi:10.1016/B978-0-12-394592-1.00003-X. PMID 23140625.
- ^ Tian X, Liu Z, Niu B; et al. (2011). "E-cadherin/β-catenin complex and the epithelial barrier". J. Biomed. Biotechnol. 2011: 567305. doi:10.1155/2011/567305. PMC 3191826. PMID 22007144.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Forbes SA, Bindal N, Bamford S; et al. (January 2011). "COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer". Nucleic Acids Res. 39 (Database issue): D945–50. doi:10.1093/nar/gkq929. PMC 3013785. PMID 20952405.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Saldanha G, Ghura V, Potter L, Fletcher A (July 2004). "Nuclear β-catenin in basal cell carcinoma correlates with increased proliferation". Br. J. Dermatol. 151 (1): 157–64. doi:10.1111/j.1365-2133.2004.06048.x. PMID 15270885.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ "Wnt/β-catenin signalling in prostate cancer". Nature Reviews Urology. Jun 2012. doi:10.1038/nrurol.2012.116. PMID 22710668.
- ^ Ashraf M. Hassanein, Steven M. Glanz, Harvey P. Kessler, Thomas A. Eskin, Chen Liu (2003). "β-Catenin Is Expressed Aberrantly in Tumors Expressing Shadow Cells". Anatomic Pathology (5) 732-6. doi: 10.1309/EALEG7LD6W7167PX. PMID 14608900.
- ^ Ellison DW, Onilude OE, Lindsey JC, Lusher ME, Weston CL, Taylor RE, Pearson AD, Clifford SC (Nov 2005). "beta-Catenin status predicts a favorable outcome in childhood medulloblastoma: the United Kingdom Children's Cancer Study Group Brain Tumour Committee". J Clin Oncol. 23 (31): 7951–7. doi:10.1200/JCO.2005.01.5479. PMID 16258095.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ M Kobayashi, T Honma, Y Matsuda, Y Suzuki, R Narisawa, Y Ajioka, H Asakura (May 2000). "Nuclear translocation of beta-catenin in colorectal cancer". Br J Cancer (10) 1689–1693. doi:10.1054/bjoc.1999.1112 PMID 10817505.
- ^ Voronkov A, Krauss S (2013). "Wnt/beta-catenin signaling and small molecule inhibitors". Curr. Pharm. Des. 19 (4): 634–64. PMC 3529405. PMID 23016862.
- ^ de la Roche M, Rutherford TJ, Gupta D; et al. (2012). "An intrinsically labile α-helix abutting the BCL9-binding site of β-catenin is required for its inhibition by carnosic acid". Nat Commun. 3 (2): 680. doi:10.1038/ncomms1680. PMC 3293410. PMID 22353711.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Takada K, Zhu D, Bird GH; et al. (August 2012). "Targeted disruption of the BCL9/β-catenin complex inhibits oncogenic Wnt signaling". Sci Transl Med. 4 (148): 148ra117. doi:10.1126/scitranslmed.3003808. PMC 3631420. PMID 22914623.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Su LK, Vogelstein B, Kinzler KW (1993). "Association of the APC tumor suppressor protein with catenins". Science. 262 (5140): 1734–7. doi:10.1126/science.8259519. PMID 8259519.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b c d Kucerová D, Sloncová E, Tuhácková Z, Vojtechová M, Sovová V (2001). "Expression and interaction of different catenins in colorectal carcinoma cells". Int. J. Mol. Med. 8 (6): 695–8. PMID 11712088.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Tickenbrock L, Kössmeier K, Rehmann H, Herrmann C, Müller O (2003). "Differences between the interaction of beta-catenin with non-phosphorylated and single-mimicked phosphorylated 20-amino acid residue repeats of the APC protein". J. Mol. Biol. 327 (2): 359–67. doi:10.1016/S0022-2836(03)00144-X. PMID 12628243.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b Davies G, Jiang WG, Mason MD (2001). "The interaction between beta-catenin, GSK3beta and APC after motogen induced cell-cell dissociation, and their involvement in signal transduction pathways in prostate cancer". Int. J. Oncol. 18 (4): 843–7. PMID 11251183.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Ryo A, Nakamura M, Wulf G, Liou YC, Lu KP (2001). "Pin1 regulates turnover and subcellular localization of beta-catenin by inhibiting its interaction with APC". Nat. Cell Biol. 3 (9): 793–801. doi:10.1038/ncb0901-793. PMID 11533658.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Homma MK, Li D, Krebs EG, Yuasa Y, Homma Y (2002). "Association and regulation of casein kinase 2 activity by adenomatous polyposis coli protein". Proceedings of the National Academy of Sciences of the United States of America. 99 (9): 5959–64. doi:10.1073/pnas.092143199. PMC 122884. PMID 11972058.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Satoh K, Yanai H, Senda T, Kohu K, Nakamura T, Okumura N, Matsumine A, Kobayashi S, Toyoshima K, Akiyama T (1997). "DAP-1, a novel protein that interacts with the guanylate kinase-like domains of hDLG and PSD-95". Genes Cells. 2 (6): 415–24. doi:10.1046/j.1365-2443.1997.1310329.x. PMID 9286858.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Eklof Spink K, Fridman SG, Weis WI (2001). "Molecular mechanisms of beta-catenin recognition by adenomatous polyposis coli revealed by the structure of an APC-beta-catenin complex". EMBO J. 20 (22): 6203–12. doi:10.1093/emboj/20.22.6203. PMC 125720. PMID 11707392.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Nakamura T, Hamada F, Ishidate T, Anai K, Kawahara K, Toyoshima K, Akiyama T (1998). "Axin, an inhibitor of the Wnt signalling pathway, interacts with beta-catenin, GSK-3beta and APC and reduces the beta-catenin level". Genes Cells. 3 (6): 395–403. doi:10.1046/j.1365-2443.1998.00198.x. PMID 9734785.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Hocevar BA, Mou F, Rennolds JL, Morris SM, Cooper JA, Howe PH (2003). "Regulation of the Wnt signaling pathway by disabled-2 (Dab2)". EMBO J. 22 (12): 3084–94. doi:10.1093/emboj/cdg286. PMC 162138. PMID 12805222.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Yang F, Li X, Sharma M, Sasaki CY, Longo DL, Lim B, Sun Z (2002). "Linking beta-catenin to androgen-signaling pathway". J. Biol. Chem. 277 (13): 11336–44. doi:10.1074/jbc.M111962200. PMID 11792709.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Masiello D, Chen SY, Xu Y, Verhoeven MC, Choi E, Hollenberg AN, Balk SP (2004). "Recruitment of beta-catenin by wild-type or mutant androgen receptors correlates with ligand-stimulated growth of prostate cancer cells". Mol. Endocrinol. 18 (10): 2388–401. doi:10.1210/me.2003-0436. PMID 15256534.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Song LN, Coghlan M, Gelmann EP (2004). "Antiandrogen effects of mifepristone on coactivator and corepressor interactions with the androgen receptor". Mol. Endocrinol. 18 (1): 70–85. doi:10.1210/me.2003-0189. PMID 14593076.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Amir AL, Barua M, McKnight NC, Cheng S, Yuan X, Balk SP (2003). "A direct beta-catenin-independent interaction between androgen receptor and T cell factor 4". J. Biol. Chem. 278 (33): 30828–34. doi:10.1074/jbc.M301208200. PMID 12799378.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ a b Mulholland DJ, Read JT, Rennie PS, Cox ME, Nelson CC (2003). "Functional localization and competition between the androgen receptor and T-cell factor for nuclear beta-catenin: a means for inhibition of the Tcf signaling axis". Oncogene. 22 (36): 5602–13. doi:10.1038/sj.onc.1206802. PMID 12944908.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Pawlowski JE, Ertel JR, Allen MP, Xu M, Butler C, Wilson EM, Wierman ME (2002). "Liganded androgen receptor interaction with beta-catenin: nuclear co-localization and modulation of transcriptional activity in neuronal cells". J. Biol. Chem. 277 (23): 20702–10. doi:10.1074/jbc.M200545200. PMID 11916967.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Takemaru K, Yamaguchi S, Lee YS, Zhang Y, Carthew RW, Moon RT (2003). "Chibby, a nuclear beta-catenin-associated antagonist of the Wnt/Wingless pathway". Nature. 422 (6934): 905–9. doi:10.1038/nature01570. PMID 12712206.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Davies G, Jiang WG, Mason MD (2001). "HGF/SF modifies the interaction between its receptor c-Met, and the E-cadherin/catenin complex in prostate cancer cells". Int. J. Mol. Med. 7 (4): 385–8. PMID 11254878.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b Oyama T, Kanai Y, Ochiai A, Akimoto S, Oda T, Yanagihara K, Nagafuchi A, Tsukita S, Shibamoto S, Ito F (1994). "A truncated beta-catenin disrupts the interaction between E-cadherin and alpha-catenin: a cause of loss of intercellular adhesiveness in human cancer cell lines". Cancer Res. 54 (23): 6282–7. PMID 7954478.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Hazan RB, Kang L, Roe S, Borgen PI, Rimm DL (1997). "Vinculin is associated with the E-cadherin adhesion complex". J. Biol. Chem. 272 (51): 32448–53. doi:10.1074/jbc.272.51.32448. PMID 9405455.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Kinch MS, Clark GJ, Der CJ, Burridge K (1995). "Tyrosine phosphorylation regulates the adhesions of ras-transformed breast epithelia". J. Cell Biol. 130 (2): 461–71. doi:10.1083/jcb.130.2.461. PMC 2199929. PMID 7542250.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Jiang MC, Liao CF, Tai CC (2002). "CAS/CSE 1 stimulates E-cadhrin-dependent cell polarity in HT-29 human colon epithelial cells". Biochem. Biophys. Res. Commun. 294 (4): 900–5. doi:10.1016/S0006-291X(02)00551-X. PMID 12061792.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b c d Hazan RB, Norton L (1998). "The epidermal growth factor receptor modulates the interaction of E-cadherin with the actin cytoskeleton". J. Biol. Chem. 273 (15): 9078–84. doi:10.1074/jbc.273.15.9078. PMID 9535896.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b c Bonvini P, An WG, Rosolen A, Nguyen P, Trepel J, Garcia de Herreros A, Dunach M, Neckers LM (2001). "Geldanamycin abrogates ErbB2 association with proteasome-resistant beta-catenin in melanoma cells, increases beta-catenin-E-cadherin association, and decreases beta-catenin-sensitive transcription". Cancer Res. 61 (4): 1671–7. PMID 11245482.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b Li Y, Bharti A, Chen D, Gong J, Kufe D (1998). "Interaction of glycogen synthase kinase 3beta with the DF3/MUC1 carcinoma-associated antigen and beta-catenin". Mol. Cell. Biol. 18 (12): 7216–24. PMC 109303. PMID 9819408.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Wendeler MW, Praus M, Jung R, Hecking M, Metzig C, Gessner R (2004). "Ksp-cadherin is a functional cell-cell adhesion molecule related to LI-cadherin". Exp. Cell Res. 294 (2): 345–55. doi:10.1016/j.yexcr.2003.11.022. PMID 15023525.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b Shibata T, Chuma M, Kokubu A, Sakamoto M, Hirohashi S (2003). "EBP50, a beta-catenin-associating protein, enhances Wnt signaling and is over-expressed in hepatocellular carcinoma". Hepatology. 38 (1): 178–86. doi:10.1053/jhep.2003.50270. PMID 12830000.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b Piedra J, Miravet S, Castaño J, Pálmer HG, Heisterkamp N, García de Herreros A, Duñach M (2003). "p120 Catenin-associated Fer and Fyn tyrosine kinases regulate beta-catenin Tyr-142 phosphorylation and beta-catenin-alpha-catenin Interaction". Mol. Cell. Biol. 23 (7): 2287–97. doi:10.1128/MCB.23.7.2287-2297.2003. PMC 150740. PMID 12640114.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Kang JS, Feinleib JL, Knox S, Ketteringham MA, Krauss RS (2003). "Promyogenic members of the Ig and cadherin families associate to positively regulate differentiation". Proceedings of the National Academy of Sciences of the United States of America. 100 (7): 3989–94. doi:10.1073/pnas.0736565100. PMC 153035. PMID 12634428.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Oneyama C, Nakano H, Sharma SV (2002). "UCS15A, a novel small molecule, SH3 domain-mediated protein-protein interaction blocking drug". Oncogene. 21 (13): 2037–50. doi:10.1038/sj.onc.1205271. PMID 11960376.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Navarro P, Lozano E, Cano A (1993). "Expression of E- or P-cadherin is not sufficient to modify the morphology and the tumorigenic behavior of murine spindle carcinoma cells. Possible involvement of plakoglobin". J. Cell. Sci. 105 (4): 923–34. PMID 8227214.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b Takahashi K, Suzuki K, Tsukatani Y (1997). "Induction of tyrosine phosphorylation and association of beta-catenin with EGF receptor upon tryptic digestion of quiescent cells at confluence". Oncogene. 15 (1): 71–8. doi:10.1038/sj.onc.1201160. PMID 9233779.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b Dobrosotskaya IY, James GL (2000). "MAGI-1 interacts with beta-catenin and is associated with cell-cell adhesion structures". Biochem. Biophys. Res. Commun. 270 (3): 903–9. doi:10.1006/bbrc.2000.2471. PMID 10772923.
- ^ Geng L, Burrow CR, Li HP, Wilson PD (2000). "Modification of the composition of polycystin-1 multiprotein complexes by calcium and tyrosine phosphorylation". Biochim. Biophys. Acta. 1535 (1): 21–35. doi:10.1016/S0925-4439(00)00079-X. PMID 11113628.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Shibamoto S, Hayakawa M, Takeuchi K, Hori T, Miyazawa K, Kitamura N, Johnson KR, Wheelock MJ, Matsuyoshi N, Takeichi M (1995). "Association of p120, a tyrosine kinase substrate, with E-cadherin/catenin complexes". J. Cell Biol. 128 (5): 949–57. doi:10.1083/jcb.128.5.949. PMC 2120395. PMID 7876318.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Rao RK, Basuroy S, Rao VU, Karnaky Jr KJ, Gupta A (2002). "Tyrosine phosphorylation and dissociation of occludin-ZO-1 and E-cadherin-beta-catenin complexes from the cytoskeleton by oxidative stress". Biochem. J. 368 (Pt 2): 471–81. doi:10.1042/BJ20011804. PMC 1222996. PMID 12169098.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b Schmeiser K, Grand RJ (1999). "The fate of E- and P-cadherin during the early stages of apoptosis". Cell Death Differ. 6 (4): 377–86. doi:10.1038/sj.cdd.4400504. PMID 10381631.
- ^ Pai R, Dunlap D, Qing J, Mohtashemi I, Hotzel K, French DM (2008). "Inhibition of fibroblast growth factor 19 reduces tumor growth by modulating beta-catenin signaling". Cancer Res. 68 (13): 5086–95. doi:10.1158/0008-5472.CAN-07-2325. PMID 18593907.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Straub BK, Boda J, Kuhn C, Schnoelzer M, Korf U, Kempf T, Spring H, Hatzfeld M, Franke WW (2003). "A novel cell-cell junction system: the cortex adhaerens mosaic of lens fiber cells". J. Cell. Sci. 116 (Pt 24): 4985–95. doi:10.1242/jcs.00815. PMID 14625392.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Wahl JK, Kim YJ, Cullen JM, Johnson KR, Wheelock MJ (2003). "N-cadherin-catenin complexes form prior to cleavage of the proregion and transport to the plasma membrane". J. Biol. Chem. 278 (19): 17269–76. doi:10.1074/jbc.M211452200. PMID 12604612.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Klingelhöfer J, Troyanovsky RB, Laur OY, Troyanovsky S (2000). "Amino-terminal domain of classic cadherins determines the specificity of the adhesive interactions". J. Cell. Sci. 113 (16): 2829–36. PMID 10910767.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Kesavapany S, Lau KF, McLoughlin DM, Brownlees J, Ackerley S, Leigh PN, Shaw CE, Miller CC (2001). "p35/cdk5 binds and phosphorylates beta-catenin and regulates beta-catenin/presenilin-1 interaction". Eur. J. Neurosci. 13 (2): 241–7. doi:10.1046/j.1460-9568.2001.01376.x. PMID 11168528.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b Lamberti C, Lin KM, Yamamoto Y, Verma U, Verma IM, Byers S, Gaynor RB (2001). "Regulation of beta-catenin function by the IkappaB kinases". J. Biol. Chem. 276 (45): 42276–86. doi:10.1074/jbc.M104227200. PMID 11527961.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Roe S, Koslov ER, Rimm DL (1998). "A mutation in alpha-catenin disrupts adhesion in clone A cells without perturbing its actin and beta-catenin binding activity". Cell Adhes. Commun. 5 (4): 283–96. doi:10.3109/15419069809040298. PMID 9762469.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Aberle H, Butz S, Stappert J, Weissig H, Kemler R, Hoschuetzky H (1994). "Assembly of the cadherin-catenin complex in vitro with recombinant proteins". J. Cell. Sci. 107 (12): 3655–63. PMID 7706414.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Reuver SM, Garner CC (1998). "E-cadherin mediated cell adhesion recruits SAP97 into the cortical cytoskeleton". J. Cell. Sci. 111 (8): 1071–80. PMID 9512503.
- ^ a b Schroeder JA, Adriance MC, McConnell EJ, Thompson MC, Pockaj B, Gendler SJ (2002). "ErbB-beta-catenin complexes are associated with human infiltrating ductal breast and murine mammary tumor virus (MMTV)-Wnt-1 and MMTV-c-Neu transgenic carcinomas". J. Biol. Chem. 277 (25): 22692–8. doi:10.1074/jbc.M201975200. PMID 11950845.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Wei Y, Renard CA, Labalette C, Wu Y, Lévy L, Neuveut C, Prieur X, Flajolet M, Prigent S, Buendia MA (2003). "Identification of the LIM protein FHL2 as a coactivator of beta-catenin". J. Biol. Chem. 278 (7): 5188–94. doi:10.1074/jbc.M207216200. PMID 12466281.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Kishida S, Yamamoto H, Hino S, Ikeda S, Kishida M, Kikuchi A (1999). "DIX domains of Dvl and axin are necessary for protein interactions and their ability to regulate beta-catenin stability". Mol. Cell. Biol. 19 (6): 4414–22. PMC 104400. PMID 10330181.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Kanai Y, Ochiai A, Shibata T, Oyama T, Ushijima S, Akimoto S, Hirohashi S (1995). "c-erbB-2 gene product directly associates with beta-catenin and plakoglobin". Biochem. Biophys. Res. Commun. 208 (3): 1067–72. doi:10.1006/bbrc.1995.1443. PMID 7702605.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b Edlund S, Lee SY, Grimsby S, Zhang S, Aspenström P, Heldin CH, Landström M (2005). "Interaction between Smad7 and beta-catenin: importance for transforming growth factor beta-induced apoptosis". Mol. Cell. Biol. 25 (4): 1475–88. doi:10.1128/MCB.25.4.1475-1488.2005. PMC 548008. PMID 15684397.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Grueneberg DA, Pablo L, Hu KQ, August P, Weng Z, Papkoff J (2003). "A functional screen in human cells identifies UBF2 as an RNA polymerase II transcription factor that enhances the beta-catenin signaling pathway". Mol. Cell. Biol. 23 (11): 3936–50. doi:10.1128/MCB.23.11.3936-3950.2003. PMC 155208. PMID 12748295.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Behrens J, von Kries JP, Kühl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W (1996). "Functional interaction of beta-catenin with the transcription factor LEF-1". Nature. 382 (6592): 638–42. doi:10.1038/382638a0. PMID 8757136.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Labbé E, Letamendia A, Attisano L (2000). "Association of Smads with lymphoid enhancer binding factor 1/T cell-specific factor mediates cooperative signaling by the transforming growth factor-beta and wnt pathways". Proceedings of the National Academy of Sciences of the United States of America. 97 (15): 8358–63. doi:10.1073/pnas.150152697. PMC 26952. PMID 10890911.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Yamamoto M, Bharti A, Li Y, Kufe D (1997). "Interaction of the DF3/MUC1 breast carcinoma-associated antigen and beta-catenin in cell adhesion". J. Biol. Chem. 272 (19): 12492–4. doi:10.1074/jbc.272.19.12492. PMID 9139698.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Durum SK, Aiello FB. "Interleukin-7 induces MUC1". Cancer Biol. Ther. 2 (2): 194–5.
- ^ Schroeder JA, Adriance MC, Thompson MC, Camenisch TD, Gendler SJ (2003). "MUC1 alters beta-catenin-dependent tumor formation and promotes cellular invasion". Oncogene. 22 (9): 1324–32. doi:10.1038/sj.onc.1206291. PMID 12618757.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Li Y, Kuwahara H, Ren J, Wen G, Kufe D (2001). "The c-Src tyrosine kinase regulates signaling of the human DF3/MUC1 carcinoma-associated antigen with GSK3 beta and beta-catenin". J. Biol. Chem. 276 (9): 6061–4. doi:10.1074/jbc.C000754200. PMID 11152665.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Ren J, Li Y, Kufe D (2002). "Protein kinase C delta regulates function of the DF3/MUC1 carcinoma antigen in beta-catenin signaling". J. Biol. Chem. 277 (20): 17616–22. doi:10.1074/jbc.M200436200. PMID 11877440.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Li Y, Ren J, Yu W, Li Q, Kuwahara H, Yin L, Carraway KL, Kufe D (2001). "The epidermal growth factor receptor regulates interaction of the human DF3/MUC1 carcinoma antigen with c-Src and beta-catenin". J. Biol. Chem. 276 (38): 35239–42. doi:10.1074/jbc.C100359200. PMID 11483589.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Kennell JA, O'Leary EE, Gummow BM, Hammer GD, MacDougald OA (2003). "T-cell factor 4N (TCF-4N), a novel isoform of mouse TCF-4, synergizes with beta-catenin to coactivate C/EBPalpha and steroidogenic factor 1 transcription factors". Mol. Cell. Biol. 23 (15): 5366–75. doi:10.1128/MCB.23.15.5366-5375.2003. PMC 165725. PMID 12861022.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Mizusaki H, Kawabe K, Mukai T, Ariyoshi E, Kasahara M, Yoshioka H, Swain A, Morohashi K (2003). "Dax-1 (dosage-sensitive sex reversal-adrenal hypoplasia congenita critical region on the X chromosome, gene 1) gene transcription is regulated by wnt4 in the female developing gonad". Mol. Endocrinol. 17 (4): 507–19. doi:10.1210/me.2002-0362. PMID 12554773.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Ge X, Jin Q, Zhang F, Yan T, Zhai Q (2009). "PCAF acetylates {beta}-catenin and improves its stability". Mol. Biol. Cell. 20 (1): 419–27. doi:10.1091/mbc.E08-08-0792. PMC 2613091. PMID 18987336.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Behrens J (2008). "One hit, two outcomes for VHL-mediated tumorigenesis". Nat. Cell Biol. 10 (10): 1127–8. doi:10.1038/ncb1008-1127.
- ^ Wadham C, Gamble JR, Vadas MA, Khew-Goodall Y (2003). "The protein tyrosine phosphatase Pez is a major phosphatase of adherens junctions and dephosphorylates beta-catenin". Mol. Biol. Cell. 14 (6): 2520–9. doi:10.1091/mbc.E02-09-0577. PMC 194899. PMID 12808048.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Aicher B, Lerch MM, Müller T, Schilling J, Ullrich A (1997). "Cellular redistribution of protein tyrosine phosphatases LAR and PTPsigma by inducible proteolytic processing". J. Cell Biol. 138 (3): 681–96. doi:10.1083/jcb.138.3.681. PMC 2141638. PMID 9245795.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Fuchs M, Müller T, Lerch MM, Ullrich A (1996). "Association of human protein-tyrosine phosphatase kappa with members of the armadillo family". J. Biol. Chem. 271 (28): 16712–9. doi:10.1074/jbc.271.28.16712. PMID 8663237.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Besco JA, Hooft van Huijsduijnen R, Frostholm A, Rotter A (2006). "Intracellular substrates of brain-enriched receptor protein tyrosine phosphatase rho (RPTPrho/PTPRT)". Brain Res. 1116 (1): 50–7. doi:10.1016/j.brainres.2006.07.122. PMID 16973135.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Wang B, Kishihara K, Zhang D, Hara H, Nomoto K (February 1997). "Molecular cloning and characterization of a novel human receptor protein tyrosine phosphatase gene, hPTP-J: down-regulation of gene expression by PMA and calcium ionophore in Jurkat T lymphoma cells". Biochem. Biophys. Res. Commun. 231 (1): 77–81. doi:10.1006/bbrc.1997.6004. PMID 9070223.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Yan HX, He YQ, Dong H, Zhang P, Zeng JZ, Cao HF, Wu MC, Wang HY (December 2002). "Physical and functional interaction between receptor-like protein tyrosine phosphatase PCP-2 and beta-catenin". Biochemistry. 41 (52): 15854–60. doi:10.1021/bi026095u. PMID 12501215.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ He Y, Yan H, Dong H, Zhang P, Tang L, Qiu X, Wu M, Wang H (April 2005). "Structural basis of interaction between protein tyrosine phosphatase PCP-2 and beta-catenin". Sci. China, C, Life Sci. 48 (2): 163–7. PMID 15986889.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Tesco G, Kim TW, Diehlmann A, Beyreuther K, Tanzi RE (1998). "Abrogation of the presenilin 1/beta-catenin interaction and preservation of the heterodimeric presenilin 1 complex following caspase activation". J. Biol. Chem. 273 (51): 33909–14. doi:10.1074/jbc.273.51.33909. PMID 9852041.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Kang DE, Soriano S, Frosch MP, Collins T, Naruse S, Sisodia SS, Leibowitz G, Levine F, Koo EH (1999). "Presenilin 1 facilitates the constitutive turnover of beta-catenin: differential activity of Alzheimer's disease-linked PS1 mutants in the beta-catenin-signaling pathway". J. Neurosci. 19 (11): 4229–37. PMID 10341227.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Murayama M, Tanaka S, Palacino J, Murayama O, Honda T, Sun X, Yasutake K, Nihonmatsu N, Wolozin B, Takashima A (1998). "Direct association of presenilin-1 with beta-catenin". FEBS Lett. 433 (1–2): 73–7. doi:10.1016/S0014-5793(98)00886-2. PMID 9738936.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Puppo F, Thomé V, Lhoumeau AC, Cibois M, Gangar A, Lembo F, Belotti E, Marchetto S, Lécine P, Prébet T, Sebbagh M, Shin WS, Lee ST, Kodjabachian L, Borg JP (January 2011). "Protein tyrosine kinase 7 has a conserved role in Wnt/β-catenin canonical signalling". EMBO Rep. 12 (1): 43–9. doi:10.1038/embor.2010.185. PMID 21132015.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Bauer A, Huber O, Kemler R (1998). "Pontin52, an interaction partner of beta-catenin, binds to the TATA box binding protein". Proceedings of the National Academy of Sciences of the United States of America. 95 (25): 14787–92. doi:10.1073/pnas.95.25.14787. PMC 24527. PMID 9843967.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Barker N, Hurlstone A, Musisi H, Miles A, Bienz M, Clevers H (2001). "The chromatin remodelling factor Brg-1 interacts with beta-catenin to promote target gene activation". EMBO J. 20 (17): 4935–43. doi:10.1093/emboj/20.17.4935. PMC 125268. PMID 11532957.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Taya S, Yamamoto T, Kanai-Azuma M, Wood SA, Kaibuchi K (1999). "The deubiquitinating enzyme Fam interacts with and stabilizes beta-catenin". Genes Cells. 4 (12): 757–67. doi:10.1046/j.1365-2443.1999.00297.x. PMID 10620020.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Lewalle JM, Bajou K, Desreux J, Mareel M, Dejana E, Noël A, Foidart JM (1997). "Alteration of interendothelial adherens junctions following tumor cell-endothelial cell interaction in vitro". Exp. Cell Res. 237 (2): 347–56. doi:10.1006/excr.1997.3799. PMID 9434630.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Shasby DM, Ries DR, Shasby SS, Winter MC (2002). "Histamine stimulates phosphorylation of adherens junction proteins and alters their link to vimentin". Am. J. Physiol. Lung Cell Mol. Physiol. 282 (6): L1330-8. doi:10.1152/ajplung.00329.2001. PMID 12003790.
{{cite journal}}: CS1 maint: multiple names: authors list (link)
Further reading
- Kikuchi A (2000). "Regulation of β-catenin signaling in the Wnt pathway". Biochem. Biophys. Res. Commun. 268 (2): 243–8. doi:10.1006/bbrc.1999.1860. PMID 10679188.
- Wilson PD (2001). "Polycystin: new aspects of structure, function, and regulation". J. Am. Soc. Nephrol. 12 (4): 834–45. PMID 11274246.
- Kalluri R, Neilson EG (2004). "Epithelial-mesenchymal transition and its implications for fibrosis". J. Clin. Invest. 112 (12): 1776–84. doi:10.1172/JCI20530. PMC 297008. PMID 14679171.
- De Ferrari GV, Moon RT (2007). "The ups and downs of Wnt signaling in prevalent neurological disorders". Oncogene. 25 (57): 7545–53. doi:10.1038/sj.onc.1210064. PMID 17143299.
External links
- beta+Catenin at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- "A diverse set of proteins modulate the canonical Wnt/β-catenin signaling pathway." at cancer.gov
- "The role of β-catenin in signal transduction, cell fate determination and trans-differentiation" at nih.gov
- "Researchers Offer First Direct Proof of How Arthritis Destroys Cartilage" at rochester.edu
This article incorporates text from the United States National Library of Medicine, which is in the public domain.
















