Myosin: Difference between revisions
m →Myosin VIII: Info + refs added |
|||
| Line 59: | Line 59: | ||
====Myosin VIII==== |
====Myosin VIII==== |
||
Myosin VIII is a plant-specific myosin linked to cell division;<ref>{{cite journal |author=Reddy AS, Day IS |title=Analysis of the myosins encoded in the recently completed Arabidopsis thaliana genome sequence |journal=Genome Biol. |volume=2 |issue=7 |pages=RESEARCH0024 |year=2001 |pmid=11516337 |pmc=55321 |doi= |url=http://www.ncbi.nlm.nih.gov/pmc/articles/PMC55321/?report=abstract}}</ref> specifically, it is involved in regulating the flow of cytoplasm between cells<ref>{{cite journal |author=Baluska F, Cvrcková F, Kendrick-Jones J, Volkmann D |title=Sink plasmodesmata as gateways for phloem unloading. Myosin VIII and calreticulin as molecular determinants of sink strength? |journal=Plant Physiol. |volume=126 |issue=1 |pages=39–46 |year=2001 |month=May |pmid=11351069 |pmc=1540107 |doi= |url=http://www.ncbi.nlm.nih.gov/pubmed/11351069?dopt=Abstract}}</ref> and in the localisation of [[vesicles]] to the [[phragmoplast]].<ref>{{cite journal |author=Reichelt S, Knight AE, Hodge TP, Baluska F, Samaj J, Volkmann D, Kendrick-Jones J |title=Characterization of the unconventional myosin VIII in plant cells and its localization at the post-cytokinetic cell wall |journal=The Plant Journal |volume=19 |issue=5 |pages=555-567 |year=2002 |month=Jan |doi=10.1046/j.1365-313X.1999.00553.x |url=http://www3.interscience.wiley.com/journal/119095847/abstract}}</ref> |
|||
====Myosin IX==== |
====Myosin IX==== |
||
====Myosin X==== |
====Myosin X==== |
||
Revision as of 17:43, 8 July 2010
Myosins are a large family of motor proteins found in eukaryotic tissues. They are responsible for actin-based motility.
- "The term “myosin” was originally used to describe a group of similar, but nonidentical, ATPases found in striated and smooth muscle cells." From Pollard and Korn, 1973[1]
Following the discovery by Pollard and Korn of enzymes with myosin-like function in Acanthamoeba castellanii, a large number of divergent myosin genes have been discovered throughout eukaryotes. Thus, although myosin was originally thought to be restricted to muscle cells (hence, "myo"), there is no single "myosin" but rather a huge superfamily of genes whose protein products share the basic properties of actin binding, ATP hydrolysis (ATPase enzyme activity), and force transduction. Virtually all eukaryotic cells contain myosin isoforms. Some isoforms have specialized functions in certain cell types (such as muscle), while other isoforms are ubiquitous.
Structure and function
Domains
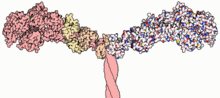
Most myosin molecules are composed of a head, neck, and tail domain.
- The head domain binds the filamentous actin, and uses ATP hydrolysis to generate force and to "walk" along the filament towards the barbed (+) end (with the exception of myosin VI, which moves towards the pointed (-) end).
- the neck domain acts as a linker and as a lever arm for transducing force generated by the catalytic motor domain. The neck domain can also serve as a binding site for myosin light chains which are distinct proteins that form part of a macromolecular complex and generally have regulatory functions.
- The tail domain generally mediates interaction with cargo molecules and/or other myosin subunits. In some cases, the tail domain may play a role in regulating motor activity.
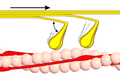
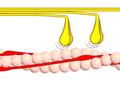
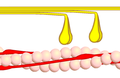
Power stroke
Multiple myosin II molecules generate force in skeletal muscle through a power stroke mechanism fuelled by the energy released from ATP hydrolysis.[2] The power stroke occurs at the release of phosphate from the myosin molecule after the ATP hydrolysis while myosin is tightly bound to actin. The effect of this release is a conformational change in the molecule that pulls against the actin. The release of the ADP molecule and binding of a new ATP molecule will release myosin from actin. ATP hydrolysis within the myosin will cause it to bind to actin again to repeat the cycle. The combined effect of the myriad power strokes causes the muscle to contract.
Nomenclature, evolution, and the family tree
The wide variety of myosin genes found throughout the eukaryotic phyla were named according to different schemes as they were discovered. The nomenclature can therefore be somewhat confusing when attempting to compare the functions of myosin proteins within and between organisms.
Skeletal muscle myosin, the most conspicuous of the myosin superfamily due to its abundance in muscle fibers, was the first to be discovered. This protein makes up part of the sarcomere and forms macromolecular filaments composed of multiple myosin subunits. Similar filament-forming myosin proteins were found in cardiac muscle, smooth muscle, and non-muscle cells. However, beginning in the 1970s researchers began to discover new myosin genes in simple eukaryotes [1] encoding proteins that acted as monomers and were therefore entitled Class I myosins. These new myosins were collectively termed "unconventional myosins" [3] and have been found in many tissues other than muscle. These new superfamily members have been grouped according to phylogenetic relationships derived from a comparison of the amino acid sequences of their head domains, with each class being assigned a Roman numeral [4][5][6][7](see phylogenetic tree). The unconventional myosins also have divergent tail domains, suggesting unique functions.[8] The now diverse array of myosins likely evolved from an ancestral precursor (see picture).
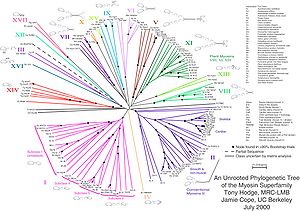
Analysis of the amino acid sequences of different myosins shows great variability among the tail domains but strong conservation of head domain sequences. Presumably this is so the myosins may interact, via their tails, with a large number of different cargoes, while the goal in each case - to move along actin filaments - remains the same and therefore requires the same machinery in the motor. For example, the human genome contains over 40 different myosin genes.
These differences in shape also determine the speed at which myosins can move along actin filaments. The hydrolysis of ATP and the subsequent release of the phosphate group causes the "power stroke," in which the "lever arm" or "neck" region of the heavy chain is dragged forward. Since the power stroke always moves the lever arm by the same angle, the length of the lever arm determines the displacement of the cargo relative to the actin filament. A longer lever arm will cause the cargo to traverse a greater distance even though the lever arm undergoes the same angular displacement - just as a person with longer legs can move farther with each individual step. The velocity of a myosin motor depends upon the rate at which it passes through a complete kinetic cycle of ATP binding to the release of ADP.
Myosin classes
Myosin I
Myosin I functions as monomer and functions in vesicle transport.[9] It has step size of 10 nm and has been implicated as being responsible for the adaptation response of the stereocilia in the inner ear.[10]
Myosin II

Myosin II (also known as conventional myosin) is the best-studied example of these properties.
- Myosin II contains two heavy chains, each about 2000 amino acids in length, which constitute the head and tail domains. Each of these heavy chains contains the N-terminal head domain, while the C-terminal tails take on a coiled-coil morphology, holding the two heavy chains together (imagine two snakes wrapped around each other, such as in a caduceus). Thus, myosin II has two heads.
- It also contains 4 light chains (2 per head), which bind the heavy chains in the "neck" region between the head and tail. These light chains are often referred to as the essential light chain and the regulatory light chain.
In muscle cells, it is myosin II that is responsible for producing the contractile force. Here, the long coiled-coil tails of the individual myosin molecules join together, forming the thick filaments of the sarcomere. The force-producing head domains stick out from the side of the thick filament, ready to walk along the adjacent actin-based thin filaments in response to the proper chemical signals.
Myosin III
Myosin III is a poorly understood member of the myosin family. It has been studied in vivo in the eyes of Drosophila, where it is thought to play a role in phototransduction.[11] A human homologue gene for myosin III, MYO3A, has been uncovered through the Human Genome Project and is expressed in the retina and cochlea.[12]
Myosin IV
Myosin IV is unconventional in that it moves to the minus end of f-actin filaments (pointed).
Myosin V
Myosin V is an unconventional myosin motor, which is functional as a dimer. It translocates (walks) along actin filaments traveling towards the barbed end (+ end) of the filaments. Myosin V was thought to be critical in vesicle movement from the center of the cell to the periphery, but has been shown to be more like a dynamic tether retaining vesicles and organelles in the actin-rich periphery of cells.
Myosin VI
Myosin VI is an unconventional myosin motor, which is functional as a dimer. It translocates (walks) along actin filaments traveling towards the pointed end of the filaments. It is the only myosin motor class known to translocate towards the pointed end. Myosin VI is thought to transport endocytic vesicles into the cell.
Myosin VII
Myosin VII is an unconventional myosin with two FERM domains in the tail region. It has a very short coiled-coiled region. Myosin VII is required for phagocytosis in Dictyostelium, spermatogenesis in C. elegans and stereocilia formation in mice and zebrafish.[13]
Myosin VIII
Myosin VIII is a plant-specific myosin linked to cell division;[14] specifically, it is involved in regulating the flow of cytoplasm between cells[15] and in the localisation of vesicles to the phragmoplast.[16]
Myosin IX
Myosin X
Myosin X is an unconventional myosin motor, which is functional as a dimer. In mammalian cells the motor is found to localize to filopodia. Myosin X walks towards the barbed ends of filaments. Some research suggests that it preferentially walks on bundles of actin, rather than single filaments. It is the first myosin motor to exhibit this behavior.
Myosin XI
Myosin XI directs the movement of organelles as plastids and mitochondria in plant cells[17]. It is responsible of the light-directed movement of chloroplasts according to light intensity and the formation of stromules interconnecting different plastids.
Myosin XII
Myosin XIII
Myosin XIV
Myosin XV
Myosin XV is a motor that is involved in the structuring and motion of stereocilia of in the inner ear. It is thought to be functional as a monomer.
Myosin XVI
Myosin XVII
Myosin XVIII
Genes in humans
Note that not all of these genes are active.
- Class I: MYO1A, MYO1B, MYO1C, MYO1D, MYO1E, MYO1F, MYO1G, MYO1H
- Class II: MYH1, MYH2, MYH3, MYH4, MYH6, MYH7, MYH7B, MYH8, MYH9, MYH10, MYH11, MYH13, MYH14, MYH15, MYH16
- Class III: MYO3A, MYO3B
- Class V: MYO5A, MYO5B, MYO5C
- Class VI: MYO6
- Class VII: MYO7A, MYO7B
- Class IX: MYO9A, MYO9B
- Class X: MYO10
- Class XV: MYO15A
- Class XVIII: MYO18A, MYO18B
Myosin light chains are distinct and have their own properties. They are not considered "myosins" but are components of the macromolecular complexes that make up the functional myosin enzymes.
Paramyosin
Paramyosin is a large 93-115kDa muscle protein that has been described in a number of diverse invertebrate phyla.[18] It is thought that invertebrate thick filaments are composted of an inner paramyosin core that is surrounded by myosin. The myosin interacts with actin resulting in fibre contraction.[19] Paramyosin is found in many different invertebrate species, for example, Brachiopoda, Sipunculidea, Nematoda, Annelida, Mollusca, Arachnida, Insecta[18]. Paramyosin is responsible for the "catch" mechanism that enables sustained contraction of muscles with very little energy expenditure, such that a clam can remain closed for extended periods.
Footnotes
- ^ a b Pollard and Korn, 1973
- ^ Tyska MJ, Warshaw DM (2002). "The myosin power stroke". Cell Motil Cytoskeleton. 51 (1): 1–15. doi:10.1002/cm.10014. PMID 11810692.
- ^ Cheney and Mooseker, 1992
- ^ Cheney et al., 1993
- ^ Goodson, 2004
- ^ Hodge and Cope, 2000
- ^ Berg et al., 2001
- ^ Oliver et al., 1999
- ^ Sutherland Macive (6/4/03). "Myosin I". Retrieved 2007-05-23.
{{cite web}}: Check date values in:|date=(help) - ^ {{cite journal | author =Batters C, Arthur CP, Lin A, Porter J, Geeves MA, Milligan RA, Molloy JE, Coluccio LM. | title = Myo1c is designed for the adaptation response in the inner ear. | journal = EMBO | year = 2004 | volume = 23| pages = 1433-1440 }
- ^ http://www.bms.ed.ac.uk/research/others/smaciver/Myosin%20III.htm
- ^ "Entrez Gene: MYO3A myosin IIIA".
- ^ Schliwa, 2003, pp 516, 518
- ^ Reddy AS, Day IS (2001). "Analysis of the myosins encoded in the recently completed Arabidopsis thaliana genome sequence". Genome Biol. 2 (7): RESEARCH0024. PMC 55321. PMID 11516337.
- ^ Baluska F, Cvrcková F, Kendrick-Jones J, Volkmann D (2001). "Sink plasmodesmata as gateways for phloem unloading. Myosin VIII and calreticulin as molecular determinants of sink strength?". Plant Physiol. 126 (1): 39–46. PMC 1540107. PMID 11351069.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Reichelt S, Knight AE, Hodge TP, Baluska F, Samaj J, Volkmann D, Kendrick-Jones J (2002). "Characterization of the unconventional myosin VIII in plant cells and its localization at the post-cytokinetic cell wall". The Plant Journal. 19 (5): 555–567. doi:10.1046/j.1365-313X.1999.00553.x.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Amir Sattarzadeh, Johanna Krahmer, Arnaud D. Germain and Maureen R. Hanson (2009). "A Myosin XI Tail Domain Homologous to the Yeast Myosin Vacuole-Binding Domain Interacts with Plastids and Stromules in Nicotiana benthamiana". Molecular Plant. 2(6): 1351–1358.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b Winkelman, L. 1976
- ^ Twarog, B. M. 1976
References
- Berg JS, Powell BC, Cheney RE (2001). "A millennial myosin census". Mol. Biol. Cell. 12 (4): 780–94. PMC 32266. PMID 11294886.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - Cheney RE, Mooseker MS (1992). "Unconventional myosins". Curr. Opin. Cell Biol. 4 (1): 27–35. doi:10.1016/0955-0674(92)90055-H. PMID 1558751.
{{cite journal}}: Unknown parameter|month=ignored (help) - Cheney RE, Riley MA, Mooseker MS (1993). "Phylogenetic analysis of the myosin superfamily". Cell Motil. Cytoskeleton. 24 (4): 215–23. doi:10.1002/cm.970240402. PMID 8477454.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Hodge T, Cope MJ (2000). "A myosin family tree". J. Cell. Sci. 113 Pt 19: 3353–4. PMID 10984423.
{{cite journal}}: Unknown parameter|month=ignored (help) - Gavin RH (2001). "Myosins in protists". Int. Rev. Cytol. 206: 97–134. doi:10.1016/S0074-7696(01)06020-X. PMID 11407764.
- Goodson HV (1994). "Molecular evolution of the myosin superfamily: application of phylogenetic techniques to cell biological questions". Soc. Gen. Physiol. Ser. 49: 141–57. PMID 7939893.
- Mooseker MS, Cheney RE (1995). "Unconventional myosins". Annu. Rev. Cell Dev. Biol. 11: 633–75. doi:10.1146/annurev.cb.11.110195.003221. PMID 8689571.
- Oliver TN, Berg JS, Cheney RE (1999). "Tails of unconventional myosins". Cell. Mol. Life Sci. 56 (3–4): 243–57. doi:10.1007/s000180050426. PMID 11212352.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - Pollard TD, Korn ED (1973). "Acanthamoeba myosin. I. Isolation from Acanthamoeba castellanii of an enzyme similar to muscle myosin". J. Biol. Chem. 248 (13): 4682–90. PMID 4268863.
{{cite journal}}: Unknown parameter|month=ignored (help) - Sellers JR (2000). "Myosins: a diverse superfamily". Biochim. Biophys. Acta. 1496 (1): 3–22. doi:10.1016/S0167-4889(00)00005-7. PMID 10722873.
{{cite journal}}: Unknown parameter|month=ignored (help) - Soldati T, Geissler H, Schwarz EC (1999). "How many is enough? Exploring the myosin repertoire in the model eukaryote Dictyostelium discoideum". Cell Biochem. Biophys. 30 (3): 389–411. doi:10.1007/BF02738121. PMID 10403058.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Twarog, B. M. (1976). Aspects of smooth muscle function in molluscan catch muscle. Current Topics in Physiology 56, 829-838.
- Winkelman, L. (1976). Comparative studies of paramyosins. Comparative Biochemistry and Physiology, Part B, Biochemistry and Molecular Biology 55, 391-397.
- Molecular Biology of the Cell. Alberts, Johnson, Lewis, Raff, Roberts, and Walter. 4th Edition. 949-952.
- Manfred Schliwa, ed. (2003). Molecular Motors. Wiley-VCH. ISBN 3-527-30594-7.
Additional images
-
Phase 1
-
Phase 2
-
Phase 3
-
Phase 4
-
Myosin-painting
-
Myosin powerstroke
See also
External links
- Myosin Video A video of a moving myosin motor protein.
- Myosins at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- The Myosin Homepage
- Animation of a moving myosin motor protein
- EC 3.6.4.1