Ligand-gated ion channel: Difference between revisions
→Cys-loop receptors: New protein nomenclature recommendations introduced |
m →Cys-loop receptors: typo |
||
| Line 68: | Line 68: | ||
=== Cys-loop receptors === |
=== Cys-loop receptors === |
||
The [[cys-loop receptors]] contain a characteristic loop formed by a disulfide bond between two [[cysteine]] residues and are subdivided into the type of ion that the corresponding channel conducts (anionic or cationic) and further into families defined by the |
The [[cys-loop receptors]] contain a characteristic loop formed by a disulfide bond between two [[cysteine]] residues and are subdivided into the type of ion that the corresponding channel conducts (anionic or cationic) and further into families defined by the engogenous ligand. |
||
'''Anionic''' |
'''Anionic''' |
||
Revision as of 12:11, 19 December 2008
| Neurotransmitter-gated ion-channel transmembrane region | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
 Ligand-gated ion channel | |||||||||||
| Identifiers | |||||||||||
| Symbol | Neur_chan_memb | ||||||||||
| Pfam | PF02932 | ||||||||||
| InterPro | IPR006029 | ||||||||||
| PROSITE | PDOC00209 | ||||||||||
| SCOP2 | 1cek / SCOPe / SUPFAM | ||||||||||
| TCDB | 1.A.9 | ||||||||||
| OPM superfamily | 14 | ||||||||||
| OPM protein | 2bg9 | ||||||||||
| |||||||||||
| Neurotransmitter-gated ion-channel ligand binding domain | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
 | |||||||||||
| Identifiers | |||||||||||
| Symbol | Neur_chan_LBD | ||||||||||
| Pfam | PF02931 | ||||||||||
| InterPro | IPR006202 | ||||||||||
| PROSITE | PDOC00209 | ||||||||||
| SCOP2 | 1lxg / SCOPe / SUPFAM | ||||||||||
| |||||||||||
Ligand-gated ion channels (LGICs), also referred to as ionotropic receptors or channel-linked receptors, are a group of transmembrane ion channels that are opened or closed in response to the binding of a chemical messenger (i.e., a ligand),[1] such as a neurotransmitter.[2]
The direct link to an ion channel, which is characteristic of ligand-gated ion channels, is contrasted with the indirect function of metabotropic receptors, which use second messengers. Ligand-gated ion channels are also different from voltage-gated ion channels (which open and close depending on membrane potential), and stretch-activated ion channels (which open and close depending on mechanical deformation of the cell membrane).[2][3]
Regulation
The ion channel is regulated by a ligand and is usually very selective to one or more ions like Na+, K+, Ca2+, or Cl-. Such receptors located at synapses convert the chemical signal of presynaptically released neurotransmitter directly and very quickly into a postsynaptic electrical signal.
Many LGICs are additionally modulated by allosteric ligands, by channel blockers, ions, or the membrane potential.
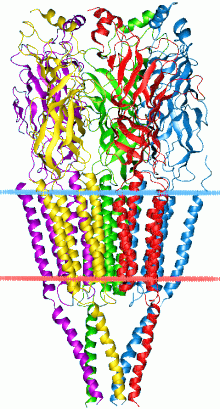
Structure
Each subunit of the pentameric channels consist of the extracellular ligand-binding domain and a transmembrane domain. Each transmembrane domain in the pentamer includes four transmembrane helixes.[4]
Example: nicotinic acetylcholine receptor
The prototypic ligand-gated ion channel is the nicotinic acetylcholine receptor. It consists of a pentamer of protein subunits, with two binding sites for acetylcholine, which, when bound, alter the receptor's configuration and cause an internal pore to open. This pore allows Na+ ions to flow down their electrochemical gradient into the cell. With a sufficient number of channels opening at once, the intracellular Na+ concentration rises to the point at which the positive charge within the cell is enough to depolarize the membrane, and an action potential is initiated.
Classification
Many important ion channels are ligand-gated, and they show a significant degree of homology at the genetic level. The Ligand-gated ion channels are classified into three superfamilies:
Cys-loop receptors
The cys-loop receptors contain a characteristic loop formed by a disulfide bond between two cysteine residues and are subdivided into the type of ion that the corresponding channel conducts (anionic or cationic) and further into families defined by the engogenous ligand.
Anionic
| Type | Class | IUPHAR-recommended protein name [5] |
Gene | Previous names |
|---|---|---|---|---|
| GABAA | alpha | α1 α2 α3 α4 α5 α6 |
GABRA1 GABRA2 GABRA3 GABRA4 GABRA5 GABRA6 |
|
| beta | β1 β2 β3 |
GABRB1 GABRB2 GABRB3 |
||
| gamma | γ1 γ2 γ3 |
GABRG1 GABRG2 GABRG3 |
||
| delta | δ | GABRD | ||
| epsilon | ε | GABRE | ||
| pi | π | GABRP | ||
| theta | θ | GABRQ | ||
| rho | ρ1 ρ2 ρ3 |
GABRR1 GABRR2 GABRR3 |
GABAC [6] | |
| Glycine (GlyR) |
alpha | α1 α2 α3 α4 |
GLRA1 GLRA2 GLRA3 GLRA4 |
|
| beta | β | GLRB |
Cationic
| Type | Class | IUPHAR-recommended protein name [5] |
Gene | Previous names |
|---|---|---|---|---|
| Serotonin (5-HT) |
5-HT3 | 5-HT3A 5-HT3B 5-HT3C 5-HT3D 5-HT3E |
HTR3A HTR3B HTR3C HTR3D HTR3E |
5-HT3A 5-HT3B 5-HT3C 5-HT3D 5-HT3E |
| Nicotinic acetylcholine (nAChR) |
alpha | α1 α2 α3 α4 α5 α6 α7 α9 α10 |
CHRNA1 CHRNA2 CHRNA3 CHRNA4 CHRNA5 CHRNA6 CHRNA7 CHRNA9 CHRNA10 |
|
| beta | β1 β2 β3 β4 |
CHRNB1 CHRNB2 CHRNB3 CHRNB4 |
||
| gamma | γ | CHRNG | ||
| delta | δ | CHRND | ||
| epsilon | ε | CHRNE | ||
| Zinc-activated channel (ZAC) |
ZAC | ZACN |
Ionotropic glutamate receptors
The glutamate receptors bind the neurotransmitter glutamate:
| Type | Class | IUPHAR-recommended protein name [5] |
Gene | Previous names |
|---|---|---|---|---|
| AMPA | GluA | GluA1 GluA2 GluA3 GluA4 |
GRIA1 GRIA2 GRIA3 GRIA4 |
GLUA1, GluR1, GluRA, GluR-A, GluR-K1, HBGR1 GLUA2, GluR2, GluRB, GluR-B, GluR-K2, HBGR2 GLUA3, GluR3, GluRC, GluR-C, GluR-K3 GLUA4, GluR4, GluRD, GluR-D |
| Kainate | GluK | GluK1 GluK2 GluK3 GluK4 GluK5 |
GRIK1 GRIK2 GRIK3 GRIK4 GRIK5 |
GLUK5, GluR5, GluR-5, EAA3 GLUK6, GluR6, GluR-6, EAA4 GLUK7, GluR7, GluR-7, EAA5 GLUK1, KA1, KA-1, EAA1 GLUK2, KA2, KA-2, EAA2 |
| NMDA | GluN | GluN1 NRL1A NRL1B |
GRIN1 GRINL1A GRINL1B |
GLUN1, NMDA-R1, NR1, GluRξ1 |
| GluN2A GluN2B GluN2C GluN2D |
GRIN2A GRIN2B GRIN2C GRIN2D |
GLUN2A, NMDA-R2A, NR2A, GluRε1 GLUN2B, NMDA-R2B, NR2B, hNR3, GluRε2 GLUN2C, NMDA-R2C, NR2C, GluRε3 GLUN2D, NMDA-R2D, NR2D, GluRε4 | ||
| GluN3A GluN3B |
GRIN3A GRIN3B |
GLUN3A, NMDA-R3A, NMDAR-L, chi-1 GLU3B, NMDA-R3B |
ATP-gated channels
ATP-gated channels open in response to binding the nucleotide ATP:
| Type | Class | IUPHAR-recommended protein name [5] |
Gene | Previous names |
|---|---|---|---|---|
| P2X | N/A | P2X1 P2X2 P2X3 P2X4 P2X5 P2X6 P2X7 |
P2RX1 P2RX2 P2RX3 P2RX4 P2RX5 P2RX6 P2RX7 |
P2X1 P2X2 P2X3 P2X4 P2X5 P2X6 P2X7 |
Clinical relevance
Ligand-gated ion channels are likely to be the major site at which anaesthetic agents and ethanol have their effects, although unequivocal evidence of this is yet to be established.[7][8] In particular, the GABA and NMDA receptors are affected by anaesthetic agents at concentrations similar to those used in clinical anaesthesia.[9]
See also
References
- ^ "ligand-gated channel" at Dorland's Medical Dictionary
- ^ a b Purves, Dale, George J. Augustine, David Fitzpatrick, William C. Hall, Anthony-Samuel LaMantia, James O. McNamara, and Leonard E. White (2008). Neuroscience. 4th ed. Sinauer Associates. pp. 156–7. ISBN 978-0-87893-697-7.
{{cite book}}: CS1 maint: multiple names: authors list (link) - ^ Connolly CN, Wafford KA (2004). "The Cys-loop superfamily of ligand-gated ion channels: the impact of receptor structure on function". Biochem. Soc. Trans. 32 (Pt3): 529–34. doi:10.1042/BST0320529. PMID 15157178.
- ^ Cascio M (2004). "Structure and function of the glycine receptor and related nicotinicoid receptors". J. Biol. Chem. 279 (19): 19383–6. doi:10.1074/jbc.R300035200. PMID 15023997.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b c d Collingridge GL, Olsen RW, Peters J, Spedding M (2008). "A nomenclature for ligand-gated ion channels". Neuropharmacology. Epub ahead of print. doi:10.1016/j.neuropharm.2008.06.063. PMID 18655795.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ A">Olsen RW, Sieghart W (2008). "International Union of Pharmacology. LXX. Subtypes of γ-aminobutyric acidA receptors: classification on the basis of subunit composition, pharmacology, and function. Update". Pharmacol. Rev. 60: 243–60. PMID 18790874.
- ^ Krasowski MD, Harrison NL (1999). "General anaesthetic actions on ligand-gated ion channels". Cell. Mol. Life Sci. 55 (10): 1278–303. doi:10.1007/s000180050371. PMID 10487207.
- ^ Dilger JP (2002). "The effects of general anaesthetics on ligand-gated ion channels". Br J Anaesth. 89 (1): 41–51. doi:10.1093/bja/aef161. PMID 12173240.
- ^ Harris RA, Mihic SJ, Dildy-Mayfield JE, Machu TK (1995). "Actions of anesthetics on ligand-gated ion channels: role of receptor subunit composition" (abstract). FASEB J. 9 (14): 1454–62. PMID 7589987.
{{cite journal}}: CS1 maint: multiple names: authors list (link)
External links
- "Revised Recommendations for Nomenclature of Ligand-Gated Ion Channels". IUPHAR Database of Receptors and Ion Channels. International Union of Basic and Clinical Pharmacology.
{{cite web}}: Cite has empty unknown parameter:|coauthors=(help) - Ligand-Gated Ion Channel database at European Bioinformatics Institute. Verified availability April 11, 2007.
Further reading
- Collingridge GL, Olsen RW, Peters J, Spedding M (2008). "A nomenclature for ligand-gated ion channels". Neuropharmacology. Epub ahead of print. PMID 18655795.
{{cite journal}}: Text "doi:10.1016/j.neuropharm.2008.06.063" ignored (help)CS1 maint: multiple names: authors list (link)
