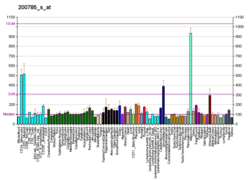
LRP1
Low density lipoprotein receptor-related protein 1 (LRP1), also known as alpha-2-macroglobulin receptor (A2MR), apolipoprotein E receptor (APOER) or cluster of differentiation 91 (CD91), is a protein forming a receptor found in the plasma membrane of cells involved in receptor-mediated endocytosis. In humans, the LRP1 protein is encoded by the LRP1 gene.[5][6][7] LRP1 is also a key signalling protein and, thus, involved in various biological processes, such as lipoprotein metabolism and cell motility, and diseases, such as neurodegenerative diseases, atherosclerosis, and cancer.[8][9]
Structure
[edit]The LRP1 gene encodes a 600 kDa precursor protein that is processed by furin in the trans-Golgi complex, resulting in a 515 kDa alpha-chain and an 85 kDa beta-chain associated noncovalently.[8][10][11] As a member of the LDLR family, LRP1 contains cysteine-rich complement-type repeats, EGF (gene) repeats, β-propeller domains, a transmembrane domain, and a cytoplasmic domain.[9] The extracellular domain of LRP1 is the alpha-chain, which comprises four ligand-binding domains (numbered I-IV) containing two, eight, ten, and eleven cysteine-rich complement-type repeats, respectively.[8][9][10][11] These repeats bind extracellular matrix proteins, growth factors, proteases, protease inhibitor complexes, and other proteins involved in lipoprotein metabolism.[8][9] Of the four domains, II and IV bind the majority of the protein's ligands.[11] The EGF repeats and β-propeller domains serve to release ligands in low pH conditions, such as inside endosomes, with the β-propeller postulated to displace the ligand at the ligand binding repeats.[9] The transmembrane domain is the β-chain, which contains a 100-residue cytoplasmic tail. This tail contains two NPxY motifs that are responsible for the protein's function in endocytosis and signal transduction.[8]
Function
[edit]LRP1 is a member of the LDLR family and ubiquitously expressed in multiple tissues, though it is most abundant in vascular smooth muscle cells (SMCs), hepatocytes, and neurons.[8][9] LRP1 plays a key role in intracellular signaling and endocytosis, which implicates it in many cellular and biological processes, including lipid and lipoprotein metabolism, protease degradation, platelet derived growth factor receptor regulation, integrin maturation and recycling, regulation of vascular tone, regulation of blood brain barrier permeability, cell growth, cell migration, inflammation, and apoptosis, as well as diseases such as neurodegenerative diseases, atherosclerosis, and cancer.[7][8][9][10][11] To elaborate, LRP1 mainly contributes to regulate protein activity by binding target proteins as a co-receptor, in conjunction with integral membrane proteins or adaptor proteins like uPA, to the lysosome for degradation.[9][10][11] In lipoprotein metabolism, the interaction between LRP1 and APOE stimulates a signaling pathway that leads to elevated intracellular cAMP levels, increased protein kinase A activity, inhibited SMC migration, and ultimately, protection against vascular disease.[9] While membrane-bound LRP1 performs endocytic clearance of proteases and inhibitors, proteolytic cleavage of its ectodomain allows the free LRP1 to compete with the membrane-bound form and prevent their clearance.[8] Several sheddases have been implicated in the proteolytic cleavage of LRP1 such as ADAM10,[12] ADAM12,[13] ADAM17[14] and MT1-MMP.[13] LRP1 is also continuously endocytosed from the membrane and recycled back to the cell surface.[9] Though the role of LRP1 in apoptosis is unclear, it is required for tPA to bind LRP1 in order to trigger the ERK1/2 signal cascade and promote cell survival.[15]
Clinical significance
[edit]Alzheimer's disease
[edit]Neurons require cholesterol to function. Cholesterol is imported into the neuron by apolipoprotein E (apoE) via LRP1 receptors on the cell surface. It has been theorized that a causal factor in Alzheimer's is the decrease of LRP1 mediated by the metabolism of the amyloid precursor protein, leading to decreased neuronal cholesterol and increased amyloid beta.[16]
LRP1 is also implicated in the effective clearance of Aβ from the brain to the periphery across the blood-brain barrier.[17][18] LRP1 mediates pathways that interact with astrocytes and pericytes, which are associated with the blood-brain barrier. In support of this, LRP1 expression is reduced in endothelial cells as a result of normal aging and Alzheimer's disease in humans and animal models of the disease.[19][20] This clearance mechanism is modulated by the apoE isoforms, with the presence of the apoE4 isoform resulting in reduced transcytosis of Aβ in in vitro models of the blood-brain barrier.[21] The reduced clearance appears to be, at least in part, as a result of an increase in the ectodomain shedding of LRP1 by sheddases, resulting in the formation of soluble LRP1 which is no longer able to transcytose the Aβ peptides.[22]
In addition, over-accumulation of copper in the brain is associated with reduced LRP1 mediated clearance of amyloid beta across the blood brain barrier. This defective clearance may contribute to the buildup of neurotoxic amyloid-beta that is thought to contribute to Alzheimer's disease.[23]
Cardiovascular disease
[edit]Studies have elucidated different roles for LRP1 in cellular processes relevant for cardiovascular disease. Atherosclerosis is the primary cause of cardiovascular disease such as stroke and heart attacks. In the liver LRP1 is important for the removal of atherogenic lipoproteins (Chylomicron remnants, VLDL) and other proatherogenic ligands from the circulation.[24][25] LRP1 has a cholesterol-independent role in atherosclerosis by modulating the activity and cellular localization of the PDGFR-β in vascular smooth muscle cells.[26][27] Finally, LRP1 in macrophages has an effect on atherosclerosis through the modulation of the extracellular matrix and inflammatory responses.[28][29]
Cancer
[edit]LRP1 is involved in tumorigenesis, and is proposed to be a tumor suppressor. Notably, LRP1 functions in clearing proteases such as plasmin, urokinase-type plasminogen activator, and metalloproteinases, which contributes to prevention of cancer invasion, while its absence is linked to increased cancer invasion. However, the exact mechanisms require further study, as other studies have shown that LRP1 may also promote cancer invasion. One possible mechanism for the inhibitory function of LRP1 in cancer involves the LRP1-dependent endocytosis of 2′-hydroxycinnamaldehyde (HCA), resulting in decreased pepsin levels and, consequently, tumor progression.[9] Alternatively, LRP1 may regulate focal adhesion disassembly of cancer cells through the ERK and JNK pathways to aid invasion.[8] Moreover, LRP1 interacts with PAI-1 to recruit mast cells (MCs) and induce their degranulation, resulting in the release of MC mediators, activation of an inflammatory response, and development of glioma.[10]
Interactions
[edit]LRP1 has been shown to interact with:
- A2-Macroglobulin,[9]
- β-amyloid precursor protein,[9]
- APBB1,[30]
- APOE,[9][31][32]
- Aprotinin,[9]
- C1S/C1q inhibitor,[9]
- CALR,[9][33]
- CD44,[8]
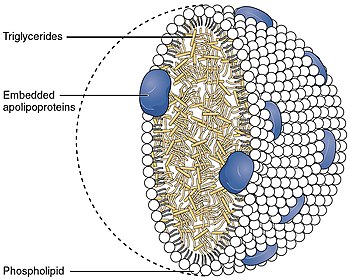
- Chylomicron,[9]
- Circumsporozoite protein,[9]
- Collectin,[9]
- Complement C3,[9]
- CTGF,[9]
- DLG4,[34]
- Elastase,[9]
- Factor IXa,[9]
- Factor VIIa,[9]
- Fibronectin,[9]
- Gentamicin,[9]
- GIPC1,[34]
- Heat shock proteins: gp96, hsp70, hsp90,[35]
- heparin cofactor II,[9]
- Hepatic lipase,[9]
- ITGB1BP1,[34]
- Lactoferrin,[9]
- Lipoprotein lipase,[9]
- LPL,[36][37][38]
- MAPK8IP1,[34]
- MAPK8IP2,[34]
- Midkine,[9]
- MMP13,[8][9]
- MMP2,[8]
- MMP9,[8][9]
- Neuroserpin,[9]
- Nexin-1,[9]
- NOS1AP,[34]
- PAI 2,[8]
- PAI-1,[8][10]
- PDGF,[9]
- tPA,[8][9]
- uPA,[8][9]
- Polymyxin B,[9]
- Protein C inhibitor,[9]
- Pseudomonas exotoxin A,[9]
- RAP,[9]
- Ricin A,[9]
- SHC1,[39][40] and
- Sphingolipid activator protein,[9]
- SYNJ2BP.[34]
- Tat,[9]
- Thrombin,[9]
- THBS1,[9][41][42][43]
- Thrombospondin 2,[9]
- TIMP1,[8]
- TIMP2,[8]
- TIMP3,[8]
- Tissue factor pathway inhibitor,[9]
- PLAT,[44][45]
- Transforming growth factor-β,[9]
- PLAUR,[46]
- VLDL,[9]
Interactive pathway map
[edit]Click on genes, proteins and metabolites below to link to respective articles. [§ 1]
- ^ The interactive pathway map can be edited at WikiPathways: "Statin_Pathway_WP430".
See also
[edit]References
[edit]- ^ a b c GRCh38: Ensembl release 89: ENSG00000123384 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000040249 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ Herz J, Hamann U, Rogne S, Myklebost O, Gausepohl H, Stanley KK (Dec 1988). "Surface location and high affinity for calcium of a 500-kd liver membrane protein closely related to the LDL-receptor suggest a physiological role as lipoprotein receptor". The EMBO Journal. 7 (13): 4119–27. doi:10.1002/j.1460-2075.1988.tb03306.x. PMC 455121. PMID 3266596.
- ^ Myklebost O, Arheden K, Rogne S, Geurts van Kessel A, Mandahl N, Herz J, et al. (Jul 1989). "The gene for the human putative apoE receptor is on chromosome 12 in the segment q13-14". Genomics. 5 (1): 65–9. doi:10.1016/0888-7543(89)90087-6. PMID 2548950.
- ^ a b "Entrez Gene: LRP1 low density lipoprotein receptor-related protein 1".
- ^ a b c d e f g h i j k l m n o p q r s t Etique N, Verzeaux L, Dedieu S, Emonard H (2013). "LRP-1: a checkpoint for the extracellular matrix proteolysis". BioMed Research International. 2013: 152163. doi:10.1155/2013/152163. PMC 3723059. PMID 23936774.
- ^ a b c d e f g h i j k l m n o p q r s t u v w x y z aa ab ac ad ae af ag ah ai aj ak al am an ao ap aq ar as at au av aw ax ay az Lillis AP, Mikhailenko I, Strickland DK (Aug 2005). "Beyond endocytosis: LRP function in cell migration, proliferation and vascular permeability". Journal of Thrombosis and Haemostasis. 3 (8): 1884–93. doi:10.1111/j.1538-7836.2005.01371.x. PMID 16102056. S2CID 20991690.
- ^ a b c d e f Roy A, Coum A, Marinescu VD, Põlajeva J, Smits A, Nelander S, et al. (Jun 2015). "Glioma-derived plasminogen activator inhibitor-1 (PAI-1) regulates the recruitment of LRP1 positive mast cells". Oncotarget. 6 (27): 23647–61. doi:10.18632/oncotarget.4640. PMC 4695142. PMID 26164207.
- ^ a b c d e Kang HS, Kim J, Lee HJ, Kwon BM, Lee DK, Hong SH (Aug 2014). "LRP1-dependent pepsin clearance induced by 2'-hydroxycinnamaldehyde attenuates breast cancer cell invasion". The International Journal of Biochemistry & Cell Biology. 53: 15–23. doi:10.1016/j.biocel.2014.04.021. PMID 24796846.
- ^ Shackleton B, Crawford F, Bachmeier C (August 2016). "Inhibition of ADAM10 promotes the clearance of Aβ across the BBB by reducing LRP1 ectodomain shedding". Fluids and Barriers of the CNS. 13 (1): 14. doi:10.1186/s12987-016-0038-x. PMC 4977753. PMID 27503326.
- ^ a b Selvais C, D'Auria L, Tyteca D, Perrot G, Lemoine P, Troeberg L, et al. (August 2011). "Cell cholesterol modulates metalloproteinase-dependent shedding of low-density lipoprotein receptor-related protein-1 (LRP-1) and clearance function". FASEB Journal. 25 (8): 2770–2781. doi:10.1096/fj.10-169508. PMC 3470721. PMID 21518850.
- ^ Liu Q, Zhang J, Tran H, Verbeek MM, Reiss K, Estus S, et al. (April 2009). "LRP1 shedding in human brain: roles of ADAM10 and ADAM17". Molecular Neurodegeneration. 4: 17. doi:10.1186/1750-1326-4-17. PMC 2672942. PMID 19371428.
- ^ Hu K, Lin L, Tan X, Yang J, Bu G, Mars WM, et al. (March 2008). "tPA protects renal interstitial fibroblasts and myofibroblasts from apoptosis". Journal of the American Society of Nephrology. 19 (3): 503–514. doi:10.1681/ASN.2007030300. PMC 2391054. PMID 18199803.
- ^ Liu Q, Zerbinatti CV, Zhang J, Hoe HS, Wang B, Cole SL, et al. (Oct 2007). "Amyloid precursor protein regulates brain apolipoprotein E and cholesterol metabolism through lipoprotein receptor LRP1". Neuron. 56 (1): 66–78. doi:10.1016/j.neuron.2007.08.008. PMC 2045076. PMID 17920016.
- ^ Deane R, Bell RD, Sagare A, Zlokovic BV (March 2009). "Clearance of amyloid-beta peptide across the blood-brain barrier: implication for therapies in Alzheimer's disease". CNS & Neurological Disorders Drug Targets. 8 (1): 16–30. doi:10.2174/187152709787601867. PMC 2872930. PMID 19275634.
- ^ Storck SE, Meister S, Nahrath J, Meißner JN, Schubert N, Di Spiezio A, et al. (January 2016). "Endothelial LRP1 transports amyloid-β(1-42) across the blood-brain barrier". The Journal of Clinical Investigation. 126 (1): 123–136. doi:10.1172/JCI81108. PMC 4701557. PMID 26619118.
- ^ Kang DE, Pietrzik CU, Baum L, Chevallier N, Merriam DE, Kounnas MZ, et al. (November 2000). "Modulation of amyloid beta-protein clearance and Alzheimer's disease susceptibility by the LDL receptor-related protein pathway". The Journal of Clinical Investigation. 106 (9): 1159–1166. doi:10.1172/JCI11013. PMC 301422. PMID 11067868.
- ^ Shibata M, Yamada S, Kumar SR, Calero M, Bading J, Frangione B, et al. (December 2000). "Clearance of Alzheimer's amyloid-ss(1-40) peptide from brain by LDL receptor-related protein-1 at the blood-brain barrier". The Journal of Clinical Investigation. 106 (12): 1489–1499. doi:10.1172/JCI10498. PMC 387254. PMID 11120756.
- ^ Bachmeier C, Paris D, Beaulieu-Abdelahad D, Mouzon B, Mullan M, Crawford F (2013-01-01). "A multifaceted role for apoE in the clearance of beta-amyloid across the blood-brain barrier". Neuro-Degenerative Diseases. 11 (1): 13–21. doi:10.1159/000337231. PMID 22572854. S2CID 30189180.
- ^ Bachmeier C, Shackleton B, Ojo J, Paris D, Mullan M, Crawford F (December 2014). "Apolipoprotein E isoform-specific effects on lipoprotein receptor processing". Neuromolecular Medicine. 16 (4): 686–696. doi:10.1007/s12017-014-8318-6. PMC 4280344. PMID 25015123.
- ^ Singh I, Sagare AP, Coma M, Perlmutter D, Gelein R, Bell RD, et al. (Sep 2013). "Low levels of copper disrupt brain amyloid-β homeostasis by altering its production and clearance". Proceedings of the National Academy of Sciences of the United States of America. 110 (36): 14771–6. Bibcode:2013PNAS..11014771S. doi:10.1073/pnas.1302212110. PMC 3767519. PMID 23959870.
- ^ Gordts PL, Reekmans S, Lauwers A, Van Dongen A, Verbeek L, Roebroek AJ (Sep 2009). "Inactivation of the LRP1 intracellular NPxYxxL motif in LDLR-deficient mice enhances postprandial dyslipidemia and atherosclerosis". Arteriosclerosis, Thrombosis, and Vascular Biology. 29 (9): 1258–64. doi:10.1161/ATVBAHA.109.192211. PMID 19667105.
- ^ Rohlmann A, Gotthardt M, Hammer RE, Herz J (Feb 1998). "Inducible inactivation of hepatic LRP gene by cre-mediated recombination confirms role of LRP in clearance of chylomicron remnants". The Journal of Clinical Investigation. 101 (3): 689–95. doi:10.1172/JCI1240. PMC 508614. PMID 9449704.
- ^ Boucher P, Gotthardt M, Li WP, Anderson RG, Herz J (Apr 2003). "LRP: role in vascular wall integrity and protection from atherosclerosis". Science. 300 (5617): 329–32. Bibcode:2003Sci...300..329B. doi:10.1126/science.1082095. PMID 12690199. S2CID 2070128.
- ^ Boucher P, Li WP, Matz RL, Takayama Y, Auwerx J, Anderson RG, et al. (2007). "LRP1 functions as an atheroprotective integrator of TGFbeta and PDFG signals in the vascular wall: implications for Marfan syndrome". PLOS ONE. 2 (5): e448. Bibcode:2007PLoSO...2..448B. doi:10.1371/journal.pone.0000448. PMC 1864997. PMID 17505534.

- ^ Yancey PG, Ding Y, Fan D, Blakemore JL, Zhang Y, Ding L, et al. (Jul 2011). "Low-density lipoprotein receptor-related protein 1 prevents early atherosclerosis by limiting lesional apoptosis and inflammatory Ly-6Chigh monocytosis: evidence that the effects are not apolipoprotein E dependent". Circulation. 124 (4): 454–64. doi:10.1161/CIRCULATIONAHA.111.032268. PMC 3144781. PMID 21730304.
- ^ Overton CD, Yancey PG, Major AS, Linton MF, Fazio S (Mar 2007). "Deletion of macrophage LDL receptor-related protein increases atherogenesis in the mouse". Circulation Research. 100 (5): 670–7. doi:10.1161/01.RES.0000260204.40510.aa. PMID 17303763.
- ^ Trommsdorff M, Borg JP, Margolis B, Herz J (Dec 1998). "Interaction of cytosolic adaptor proteins with neuronal apolipoprotein E receptors and the amyloid precursor protein". The Journal of Biological Chemistry. 273 (50): 33556–60. doi:10.1074/jbc.273.50.33556. PMID 9837937.
- ^ Poswa M (Mar 1977). "[Team growth by acquiring an apprentice]". Quintessenz Journal. 7 (3): 21–3. PMID 277965.
- ^ Kowal RC, Herz J, Goldstein JL, Esser V, Brown MS (Aug 1989). "Low density lipoprotein receptor-related protein mediates uptake of cholesteryl esters derived from apoprotein E-enriched lipoproteins". Proceedings of the National Academy of Sciences of the United States of America. 86 (15): 5810–4. Bibcode:1989PNAS...86.5810K. doi:10.1073/pnas.86.15.5810. PMC 297720. PMID 2762297.
- ^ Orr AW, Pedraza CE, Pallero MA, Elzie CA, Goicoechea S, Strickland DK, et al. (Jun 2003). "Low density lipoprotein receptor-related protein is a calreticulin coreceptor that signals focal adhesion disassembly". The Journal of Cell Biology. 161 (6): 1179–89. doi:10.1083/jcb.200302069. PMC 2172996. PMID 12821648.
- ^ a b c d e f g Gotthardt M, Trommsdorff M, Nevitt MF, Shelton J, Richardson JA, Stockinger W, et al. (Aug 2000). "Interactions of the low density lipoprotein receptor gene family with cytosolic adaptor and scaffold proteins suggest diverse biological functions in cellular communication and signal transduction". The Journal of Biological Chemistry. 275 (33): 25616–24. doi:10.1074/jbc.M000955200. PMID 10827173.
- ^ Basu S, Binder RJ, Ramalingam T, Srivastava PK (Mar 2001). "CD91 is a common receptor for heat shock proteins gp96, hsp90, hsp70, and calreticulin". Immunity. 14 (3): 303–13. doi:10.1016/s1074-7613(01)00111-x. PMID 11290339.
- ^ Williams SE, Inoue I, Tran H, Fry GL, Pladet MW, Iverius PH, et al. (Mar 1994). "The carboxyl-terminal domain of lipoprotein lipase binds to the low density lipoprotein receptor-related protein/alpha 2-macroglobulin receptor (LRP) and mediates binding of normal very low density lipoproteins to LRP". The Journal of Biological Chemistry. 269 (12): 8653–8. doi:10.1016/S0021-9258(17)37017-5. PMID 7510694.
- ^ Nykjaer A, Nielsen M, Lookene A, Meyer N, Røigaard H, Etzerodt M, et al. (Dec 1994). "A carboxyl-terminal fragment of lipoprotein lipase binds to the low density lipoprotein receptor-related protein and inhibits lipase-mediated uptake of lipoprotein in cells". The Journal of Biological Chemistry. 269 (50): 31747–55. doi:10.1016/S0021-9258(18)31759-9. PMID 7989348.
- ^ Chappell DA, Fry GL, Waknitz MA, Iverius PH, Williams SE, Strickland DK (Dec 1992). "The low density lipoprotein receptor-related protein/alpha 2-macroglobulin receptor binds and mediates catabolism of bovine milk lipoprotein lipase". The Journal of Biological Chemistry. 267 (36): 25764–7. doi:10.1016/S0021-9258(18)35675-8. PMID 1281473.
- ^ Barnes H, Ackermann EJ, van der Geer P (Jun 2003). "v-Src induces Shc binding to tyrosine 63 in the cytoplasmic domain of the LDL receptor-related protein 1". Oncogene. 22 (23): 3589–97. doi:10.1038/sj.onc.1206504. PMID 12789267.
- ^ Loukinova E, Ranganathan S, Kuznetsov S, Gorlatova N, Migliorini MM, Loukinov D, et al. (May 2002). "Platelet-derived growth factor (PDGF)-induced tyrosine phosphorylation of the low density lipoprotein receptor-related protein (LRP). Evidence for integrated co-receptor function between LRP and the PDGF". The Journal of Biological Chemistry. 277 (18): 15499–506. doi:10.1074/jbc.M200427200. PMID 11854294.
- ^ Wang S, Herndon ME, Ranganathan S, Godyna S, Lawler J, Argraves WS, et al. (Mar 2004). "Internalization but not binding of thrombospondin-1 to low density lipoprotein receptor-related protein-1 requires heparan sulfate proteoglycans". Journal of Cellular Biochemistry. 91 (4): 766–76. doi:10.1002/jcb.10781. PMID 14991768. S2CID 12198474.
- ^ Mikhailenko I, Krylov D, Argraves KM, Roberts DD, Liau G, Strickland DK (Mar 1997). "Cellular internalization and degradation of thrombospondin-1 is mediated by the amino-terminal heparin binding domain (HBD). High affinity interaction of dimeric HBD with the low density lipoprotein receptor-related protein". The Journal of Biological Chemistry. 272 (10): 6784–91. doi:10.1074/jbc.272.10.6784. PMID 9045712.
- ^ Godyna S, Liau G, Popa I, Stefansson S, Argraves WS (Jun 1995). "Identification of the low density lipoprotein receptor-related protein (LRP) as an endocytic receptor for thrombospondin-1". The Journal of Cell Biology. 129 (5): 1403–10. doi:10.1083/jcb.129.5.1403. PMC 2120467. PMID 7775583.
- ^ Zhuo M, Holtzman DM, Li Y, Osaka H, DeMaro J, Jacquin M, et al. (Jan 2000). "Role of tissue plasminogen activator receptor LRP in hippocampal long-term potentiation". The Journal of Neuroscience. 20 (2): 542–9. doi:10.1523/JNEUROSCI.20-02-00542.2000. PMC 6772406. PMID 10632583.
- ^ Orth K, Madison EL, Gething MJ, Sambrook JF, Herz J (Aug 1992). "Complexes of tissue-type plasminogen activator and its serpin inhibitor plasminogen-activator inhibitor type 1 are internalized by means of the low density lipoprotein receptor-related protein/alpha 2-macroglobulin receptor". Proceedings of the National Academy of Sciences of the United States of America. 89 (16): 7422–6. Bibcode:1992PNAS...89.7422O. doi:10.1073/pnas.89.16.7422. PMC 49722. PMID 1502153.
- ^ Czekay RP, Kuemmel TA, Orlando RA, Farquhar MG (May 2001). "Direct binding of occupied urokinase receptor (uPAR) to LDL receptor-related protein is required for endocytosis of uPAR and regulation of cell surface urokinase activity". Molecular Biology of the Cell. 12 (5): 1467–79. doi:10.1091/mbc.12.5.1467. PMC 34598. PMID 11359936.
Further reading
[edit]- Li Z, Dai J, Zheng H, Liu B, Caudill M (Mar 2002). "An integrated view of the roles and mechanisms of heat shock protein gp96-peptide complex in eliciting immune response". Frontiers in Bioscience. 7 (4): d731–51. doi:10.2741/A808. PMID 11861214.
- van der Geer P (May 2002). "Phosphorylation of LRP1: regulation of transport and signal transduction". Trends in Cardiovascular Medicine. 12 (4): 160–5. doi:10.1016/S1050-1738(02)00154-8. PMID 12069755.
- May P, Herz J (May 2003). "LDL receptor-related proteins in neurodevelopment". Traffic. 4 (5): 291–301. doi:10.1034/j.1600-0854.2003.00086_4_5.x. PMID 12713657. S2CID 23565545.
- Llorente-Cortés V, Badimon L (Mar 2005). "LDL receptor-related protein and the vascular wall: implications for atherothrombosis". Arteriosclerosis, Thrombosis, and Vascular Biology. 25 (3): 497–504. doi:10.1161/01.ATV.0000154280.62072.fd. PMID 15705932.
- Huang SS, Huang JS (Oct 2005). "TGF-beta control of cell proliferation". Journal of Cellular Biochemistry. 96 (3): 447–62. doi:10.1002/jcb.20558. PMID 16088940. S2CID 83711249.
- Lillis AP, Mikhailenko I, Strickland DK (Aug 2005). "Beyond endocytosis: LRP function in cell migration, proliferation and vascular permeability". Journal of Thrombosis and Haemostasis. 3 (8): 1884–93. doi:10.1111/j.1538-7836.2005.01371.x. PMID 16102056. S2CID 20991690.
External links
[edit]- CD91+Antigen at the U.S. National Library of Medicine Medical Subject Headings (MeSH)