Opsin


Animal opsins are G-protein-coupled receptors and a group of proteins made light-sensitive via a chromophore, typically retinal. When bound to retinal, opsins become retinylidene proteins, but are usually still called opsins regardless. Most prominently, they are found in photoreceptor cells of the retina. Five classical groups of opsins are involved in vision, mediating the conversion of a photon of light into an electrochemical signal, the first step in the visual transduction cascade. Another opsin found in the mammalian retina, melanopsin, is involved in circadian rhythms and pupillary reflex but not in vision. Humans have in total nine opsins. Beside vision and light perception, opsins may also sense temperature, sound, or chemicals.
Structure and function
[edit]Animal opsins detect light and are the molecules that allow us to see. Opsins are G-protein-coupled receptors (GPCRs),[1][2] which are chemoreceptors and have seven transmembrane domains forming a binding pocket for a ligand.[3][4] The ligand for opsins is the vitamin A-based chromophore 11-cis-retinal,[5][6][7][8][9] which is covalently bound to a lysine residue[10] in the seventh transmembrane domain[11][12][13] through a Schiff-base.[14][15] However, 11-cis-retinal only blocks the binding pocket and does not activate the opsin. The opsin is only activated when 11-cis-retinal absorbs a photon of light and isomerizes to all-trans-retinal,[16][17] the receptor activating form,[18][19] causing conformal changes in the opsin,[18] which activate a phototransduction cascade.[20] Thus, a chemoreceptor is converted to a light or photo(n)receptor.[21]
In the vertebrate photoreceptor cells, all-trans-retinal is released and replaced by a newly synthesized 11-cis-retinal provided from the retinal epithelial cells. Beside 11-cis-retinal (A1), 11-cis-3,4-didehydroretinal (A2) is also found in vertebrates as ligand such as in freshwater fishes.[19] A2-bound opsins have a shifted λmax and absorption spectrum compared to A1-bound opsins.[22]
Functionally conserved residues and motifs
[edit]The seven transmembrane α-helical domains in opsins are connected by three extra-cellular and three cytoplasmic loops. Along the α-helices and the loops, many amino acid residues are highly conserved between all opsin groups, indicating that they serve important functions and thus are called functionally conserved residues. Actually, insertions and deletions in the α-helices are very rare and should preferentially occur in the loops. Therefore, different G-protein-coupled receptors have different length and homologous residues may be in different positions. To make such positions comparable between different receptors, Ballesteros and Weinstein introduced a common numbering scheme for G-protein-coupled receptors.[23] The number before the period is the number of the transmembrane domain. The number after the period is set arbitrarily to 50 for the most conserved residue in that transmembrane domain among GPCRs known in 1995. For instance in the seventh transmembrane domain, the proline in the highly conserved NPxxY7.53 motif is Pro7.50, the asparagine before is then Asp7.49, and the tyrosine three residues after is then Tyr7.53.[21] Another numbering scheme is based on cattle rhodopsin. Cattle rhodopsin has 348 amino acids and is the first opsin whose amino acid sequence[24] and whose 3D-structure were determined.[12] The cattle rhodopsin numbering scheme is widespread in the opsin literature.[21] Therefore, it is useful to use both schemes.
The retinal binding lysine
[edit]Opsins without the retinal binding lysine are not light sensitive.[25][26] In cattle rhodopsin, this lysine is the 296th amino acid[12][24] and thus according to both numbering schemes Lys2967.43. It is well conserved among opsins, so well conserved that sequences without it were not even considered opsins and thus excluded from large scale phylogenetic reconstructions.[27][28] Even so, most opsins have Lys2967.43, some have lost it during evolution: In the nemopsins from nematodes, Lys2967.43 is replaced by Arginine.[29][21] In the astropsins from sea urchins[30][21] and in the gluopsins, Lys2967.43 is replaced by glutamic acid.[21] A nemopsin is expressed in chemosensory cells in Caenorhabditis elegans. Therefore, the nemopsins are thought to be chemoreceptors.[29] The gluopsins are found in insects such as beetles, scorpionflies, dragonflies, and butterflies and moths including model organisms such as the silk moth and the tobacco hawk moth. However, the gluopsins have no known function.[21]
Such function does not need to be light detection, as some opsins are also involved in thermosensation,[31] mechanoreception such as hearing[32] detecting phospholipids, chemosensation, and other functions.[33][34] In particular, the Drosophila rhabdomeric opsins (rhabopsins, r-opsins) Rh1, Rh4, and Rh7 function not only as photoreceptors, but also as chemoreceptors for aristolochic acid. These opsins still have Lys2967.43 like other opsins. However, if this lysine is replaced by an arginine in Rh1, then Rh1 loses light sensitivity but still responds to aristolochic acid. Thus, Lys2967.43 is not needed for Rh1 to function as chemoreceptor.[26] Also the Drosophila rhabopsins Rh1 and Rh6 are involved in mechanoreception, again for mechanoreception Lys2967.43 is not needed, but needed for proper function in the photoreceptor cells.[25]
Beside these functions, an opsin without Lys2967.43, such as a gluopsin, could still be light sensitive, since in cattle rhodopsin, the retinal binding lysine can be shifted from position 296 to other positions, even into other transmembrane domains, without changing light sensitivity.[35]
-
Most known opsins have the retinal binding lysine except some among the tetraopins, The outgroup contains other G protein-coupled receptors.
-
Most tetraopsins have also the retinal binding lysine except some of the chromopsins, which are highlighted by the frame and expanded in the next image. The outgroup contains other G protein-coupled receptors including the other opsins.
-
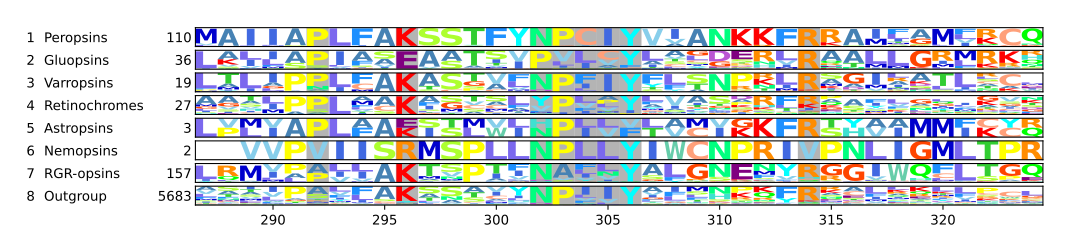
Most chromopsins have also the retinal binding lysine except the nemopsins, where it is replaced by argenine (R), and the gluopsins, where it is replaced by glutamic acid (E). The astropsins, the nemopsins and the gluopsins are highlighted by the frames. The outgroup contains other G protein-coupled receptors including the other opsins.
In the phylogeny above, each clade contains sequences from opsins and other G protein-coupled receptors. The number of sequences and two pie charts are shown next to the clade. The first pie chart shows the percentage of a certain amino acid at the position in the sequences corresponding Lys2967.43 in cattle rhodopsin. The amino acids are color-coded. The colors are red for lysine (K), purple for glutamic acid (E), orange for argenine (R), dark and mid-gray for other amino acids, and light gray for sequences that have no data at that position. The second pie chart gives the taxon composition for each clade, green stands for craniates, dark green for cephalochordates, mid green for echinoderms, brown for nematodes, pale pink for annelids, dark blue for arthropods, light blue for mollusks, and purple for cnidarians. The branches to the clades have pie charts, which give support values for the branches. The values are from right to left SH-aLRT/aBayes/UFBoot. The branches are considered supported when SH-aLRT ≥ 80%, aBayes ≥ 0.95, and UFBoot ≥ 95%. If a support value is above its threshold the pie chart is black otherwise gray.[21]
The NPxxY motif
[edit]The NPxxY7.53 motif is well-conserved among opsins and G-protein-coupled receptors. This motif is important for G-protein binding and receptor activation.[21] For instance, if it is mutated to DPxxY7.53 (Asn7.49 → Asp7.49) in the human m3 muscarinic receptor, activation is not affected, but it is abolished if it is mutated to APxxY7.53 (Asn7.49 → Ala7.49).[36] Such a mutation to APxxY7.53 (Asn7.49 → Ala7.49) reduces the G-protein activation of cattle rhodopsin to 45% compared to wild type. Also in cattle rhodopsin, if the motif is mutated to NPxxA7.53 (Tyr7.53 → Ala7.53), cattle rhodopsin does not activate the G-protein.[37] Such a mutation also reduces the activation of the vasopressin V2 receptor. In fact in G-protein-coupled receptors, only loss of function disease mutations are known for Tyr7.53.[38]
Also mutations of Pro7.50 influence G-protein activation, if the motif is mutated to NAxxY7.53 (Pro7.50 → Ala7.50) in the rat m3 muscarinic receptor, the receptor can still be activated but less efficiently,[39] this mutation even abolishes activation in the cholecystokinin B receptor completely.[40] In fact, the RGR-opsins have NAxxY7.53 and retinochromes have VPxxY7.53 for annelids or YPxxY7.53 for mollusks, natively. Both RGR-opsins and retinochromes, belong to the chromopsins.[21] RGR-opsins[41] and retinochromes[42] also bind unlike most opsins all-trans-retinal in the dark and convert it to 11-cis-retinal when illuminated. Therefore, RGR-opsins and retinochromes are thought to neither signal nor activate a phototransduction cascade but to work as photoisomerases to produce 11-cis-retinal for other opsins.[43][44] This view is considered established in the opsin literature,[34][45][43][46][47] even so it has not been shown, conclusively.[21] In fact, the human MT2 melatonin receptor signals via a G-protein and has an NAxxY7.53 motif natively. If this motif is mutated to NPxxY7.53 (Ala7.50 → Pro7.50), the receptor cannot be activated, but can be rescued partially if the motif is mutated to NVxxY7.53 (Ala7.50 → Val7.50).[48] Furthermore, when the motif is mutated to NAxxY7.53 (Pro7.50 → Ala7.50) in cattle rhodopsin, the mutant has 141% of wild type activity.[37] This evidence shows that a GPCR does not need a standard NPxxY7.53 motif for signaling.[21]
Other residues and motifs
[edit]Cys138 and Cys110 form a highly conserved disulfide bridge. Glu113 serves as the counterion, stabilizing the protonation of the Schiff linkage between Lys296 and the ligand retinal. The Glu134-Arg135-Tyr136 is another highly conserved motif, involved in the propagation of the transduction signal once a photon has been absorbed.
Spectral tuning sites
[edit]Certain amino acid residues, termed spectral tuning sites, have a strong effect on λmax values. Using site-directed mutagenesis, it is possible to selectively mutate these residues and investigate the resulting changes in light absorption properties of the opsin. It is important to differentiate spectral tuning sites, residues that affect the wavelength at which the opsin absorbs light, from functionally conserved sites, residues important for the proper functioning of the opsin. They are not mutually exclusive, but, for practical reasons, it is easier to investigate spectral tuning sites that do not affect opsin functionality. For a comprehensive review of spectral tuning sites see Yokoyama[49] and Deeb.[50] The impact of spectral tuning sites on λmax differs between different opsin groups and between opsin groups of different species.
Opsins in the human eye, brain, and skin
[edit]| Abbr. | Name | λmax | Color | Eye | Brain | Skin | Chromosomal location[44] |
|---|---|---|---|---|---|---|---|
| OPN1LW | L-cone (red-cone) opsin | 557 nm | Yellow | Cone | — | — | Xq28[44] |
| OPN1MW | M-cone (green-cone) opsin | 527 nm | Green | Cone | — | — | Xq28[44] |
| OPN1SW | S-cone (blue-cone) opsin | 420 nm | Violet | Cone | — | Melanocytes, keratinocytes[51] | 7q32.1[44] |
| OPN2 (RHO) | Rhodopsin | 505 nm | Blue–green | Rod | — | Melanocytes, keratinocytes[51] | 3q22.1[44] |
| OPN3 | Encephalopsin, panopsin | S-M | Blue–green | Rod, cone, OPL, IPL, GCL[52] | Cerebral cortex, cerebellum, striatum, thalamus, hypothalamus[53][54] | Melanocytes, keratinocytes[51] | 1q43[44] |
| OPN4 | Melanopsin | 480 nm[55] | Sky blue | ipRGC[55] | — | — | 10q23.2[44] |
| OPN5 | Neuropsin | 380 nm[56] | Ultraviolet[56] | Neural retina, RPE[57] | Anterior hypothalamus[58] | Melanocytes, keratinocytes[51] | 6p12.3[44] |
| RRH | Peropsin | RPE cells - microvilli | — | — | 4q25[44] | ||
| RGR | Retinal G protein coupled receptor | RPE cells | — | — | 10q23.1[44] |
RPE, retinal pigment epithelium; ipRGC, intrinsically photosensitive retinal ganglion cells; OPL, outer plexiform layer; IPL, inner plexiform layer; GCL, ganglion cell layer
Cuttlefish
[edit]Cuttlefish and octopuses contain opsin in their skin as part of the chromophores. The opsin is part of the sensing network detecting the colour and shape of the cuttlefish's surroundings.[59][60][61]
Phylogeny
[edit]Animal opsins (also known as type 2 opsins) are members of the seven-transmembrane-domain proteins of the G protein-coupled receptor (GPCR) superfamily.[1][2]
Animal opsins fall phylogenetically into five groups: The ciliary opsins (cilopsins, c-opsins), the rhabdomeric opsins (r-opsins, rhabopsins), the xenopsins, the nessopsins, and the tetraopsins. Four of these subclades occur in Bilateria (all but the nessopsins).[21][28] However, the bilaterian clades constitute a paraphyletic taxon without the opsins from the cnidarians.[21][28][27][62] The nessopsins are also known as anthozoan opsins II[63] or simply as the cnidarian opsins.[64] The tetraopsins are also known as RGR/Go[65] or Group 4 opsins[27] and contain three subgroups: the neuropsins, the Go-opsins, and the chromopsins.[21][28][64] The chromopsins have seven subgroups: the RGR-opsins, the retinochromes, the peropsins, the varropsins, the astropsins, the nemopsins, and the gluopsins.[21]
Animal visual opsins are traditionally classified as either ciliary or rhabdomeric. Ciliary opsins, found in vertebrates and cnidarians, attach to ciliary structures such as rods and cones. Rhabdomeric opsins are attached to light-gathering organelles called rhabdomeres. This classification cuts across phylogenetic categories (clades) so that both the terms "ciliary" and "rhabdomeric" can be ambiguous. Here, "C-opsins (ciliary)" refers to a clade found exclusively in Bilateria and excludes cnidarian ciliary opsins such as those found in the box jellyfish. Similarly, "R-opsin (rhabdomeric)" includes melanopsin even though it does not occur on rhabdomeres in vertebrates.[27]
Ciliary opsins
[edit]Ciliary opsins (cilopsins, c-opsins) are expressed in ciliary photoreceptor cells, and include the vertebrate visual opsins and encephalopsins.[66] They convert light signals to nerve impulses via cyclic nucleotide gated ion channels, which work by increasing the charge differential across the cell membrane (i.e. hyperpolarization.[67])
Vertebrate visual opsins
[edit]Vertebrate visual opsins are a subclass of ciliary opsins that express in the vertebrate retina and mediate vision. They are further subdivided into:
- Photopsins - those responsible for photopic vision (daylight), which are expressed in cone cells; hence also cone opsins. Photopsins are further subdivided according to their spectral sensitivity, namely the wavelength at which the highest light absorption is observed (λmax). Vertebrates generally have four (SWS1, SWS2, RH2, LWS) classes of photopsins.[68][69] Mammals lost Rh2 and SWS2 classes during the nocturnal bottleneck, so are generally dichromatic. Primate ancestors later developed two distinct LWS opsins (LWS and MWS), leaving humans with 3 photopsins in 2 classes: SWS1 (OPN1SW) and two forms of LWS (OPN1LW, OPN1MW).
- Scotopsins - those responsible for scotopic vision (dim light), which are expressed in rod cells; hence also rod opsins.[66] The most common form of scotopsin is rhodopsin, thus usually denoted Rh1.[70]
Extraretinal (or extra-ocular) Rhodopsin-Like Opsins (Exo-Rh)
[edit]These pineal opsins, found in the Actinopterygii (ray-finned fish) apparently arose as a result of gene duplication from Rh1 (rhodopsin). These opsins appear to serve functions similar to those of pinopsin found in birds and reptiles.[71] [72]
Pinopsins
[edit]The first Pineal Opsin (Pinopsin) was found in the chicken pineal gland. It is a blue sensitive opsin (λmax = 470 nm).[73][74]
Pineal opsins have a wide range of expression in the brain, most notably in the pineal region.
Vertebrate Ancient (VA) opsin
[edit]Vertebrate Ancient (VA) opsin has three isoforms VA short (VAS), VA medium (VAM), and VA long (VAL). It is expressed in the inner retina, within the horizontal and amacrine cells, as well as the pineal organ and habenular region of the brain.[75] It is sensitive to approximately 500 nm [14], found in most vertebrate classes, but not in mammals.[76]
Parapinopsins
[edit]The first parapinopsin (PP) was found in the parapineal organ of the catfish.[77] The parapinopsin of lamprey is a UV-sensitive opsin (λmax = 370 nm).[78] The teleosts have two groups of parapinopsins, one is sensitive to UV (λmax = 360-370 nm), the other is sensitive to blue (λmax = 460-480 nm) light.[79]
Parietopsins
[edit]The first parietopsin was found in the photoreceptor cells of the lizard parietal eye. The lizard parietopsin is green-sensitive (λmax = 522 nm), and despite it is a c-opsin, like the vertebrate visual opsins, it does not induce hyperpolarization via a Gt-protein, but induces depolarization via a Go-protein.[80][81]
Encephalopsin or Panopsin
[edit]The panopsins are found in many tissues (skin,[51] brain,[53][82] testes,[53] heart, liver,[82] kidney, skeletal muscle, lung, pancreas and retina[82]). They were originally found in the human and mouse brain and thus called encephalopsin.[53]
The first invertebrate panopsin was found in the ciliary photoreceptor cells of the annelid Platynereis dumerilii and is called c(iliary)-opsin.[83] This c-opsin is UV-sensitive (λmax = 383 nm) and can be tuned by 125 nm at a single amino-acid (range λmax = 377 - 502 nm).[84] Thus, not unsurprisingly, a second but cyan sensitive c-opsin (λmax = 490 nm) exists in Platynereis dumerilii.[85] The first c-opsin mediates in the larva UV induced gravitaxis. The gravitaxis forms with phototaxis a ratio-chromatic depth-gauge.[86] In different depths, the light in water is composed of different wavelengths: First the red (> 600 nm) and the UV and violet (< 420 nm) wavelengths disappear. The higher the depth the narrower the spectrum so that only cyan light (480 nm) is left.[87] Thus, the larvae can determine their depth by color. The color unlike brightness stays almost constant independent of time of day or the weather, for instance if it is cloudy.[88][89]
Panopsins are also expressed in the brains of some insects.[66] The panopsins of mosquito and pufferfish absorb maximally at 500 nm and 460 nm, respectively. Both activate in vitro Gi and Go proteins.[90]
The panopsins are sister to the TMT-opsins.[28][91][47][92]
Teleost Multiple Tissue (TMT) Opsin
[edit]The first TMT-opsin was found in many tissues in Teleost fish and therefore they are called Teleost Multiple Tissue (TMT) opsins.[93] TMT-opsins form three groups which are most closely related to a fourth group the panopsins, which thus are paralogous to the TMT-opsins.[28][47][91][92] TMT-opsins and panopsins also share the same introns, which confirms that they belong together.[93]
Opsins in cnidarians
[edit]Cnidaria, which include jellyfish, corals, and sea anemones, are the most basal animals to possess complex eyes. Jellyfish opsins in the rhopalia couple to Gs-proteins raising the intracellular cAMP level.[94][62] Coral opsins can couple to Gq-proteins and Gc-proteins. Gc-proteins are a subtype of G-proteins specific to cnidarians.[95] The cnidarian opsins belong to two groups the xenopsins and the nessopsins. The xenopsins contain also bilaterian opsins, while the nessopsins are restricted to the cnidarians.[21][28] However, earlier studies have found that some cnidarian opsins belong to the cilopsins, rhabopsins, and the tetraopsins of the bilaterians.[65][96][97]
Rhabdomeric opsins
[edit]Rhabdomeric opsins (rhabopsins, r-opsins) are also known as Gq-opsins, because they couple to a Gq-protein. Rhabopsins are used by molluscs and arthropods. Arthropods appear to attain colour vision in a similar fashion to the vertebrates, by using three (or more) distinct groups of opsins, distinct both in terms of phylogeny and spectral sensitivity.[66] The rhabopsin melanopsin is also expressed in vertebrates, where it regulates circadian rhythms and mediates the pupillary reflex.[66]
Unlike cilopsins, rhabopsins are associated with canonical transient receptor potential ion channels; these lead to the electric potential difference across a cell membrane being eradicated (i.e. depolarization).[67]
The identification of the crystal structure of squid rhodopsin[13] is likely to further our understanding of its function in this group.
Arthropods use different opsins in their different eye types, but at least in Limulus the opsins expressed in the lateral and the compound eyes are 99% identical and presumably diverged recently.[98]
Melanopsin
[edit]Melanopsin (OPN4) is involved in circadian rhythms, the pupillary reflex, and color correction in high-brightness situations. Phylogenetically, it is a member of the rhabdomeric opsins (rhabopsins, r-opsins) and functionally and structurally a rhabopsin, but does not occur in rhabdomeres.
Tetraopsins
[edit]The tetraopsins include the neuropsins, the Go-opsins, and the chromopsins.[21][28][64] The chromopsins consist of seven subgroups: the RGR-opsins, the retinochromes, the peropsins, the varropsins, the astropsins, the nemopsins, and the gluopsins.[21]
Neuropsins
[edit]Neuropsins are sensitive to UVA, typically at 380 nm. They are found in the brain, testes, skin, and retina of humans and rodents, as well as in the brain and retina of birds. In birds and rodents they mediate ultraviolet vision.[51][56][99] They couple to Gi-proteins.[56][99] In humans, Neuropsin is encoded by the OPN5 gene. In the human retina, its function is unknown. In the mouse, it photo-entrains the retina and cornea at least ex vivo.[100]
Go-opsins
[edit]Go-opsins are absent from higher vertebrates[27] and ecdysozoans.[101] They are found in the ciliary photoreceptor cells of the scallop eye[102] and the basal chordate amphioxus.[103] In Platynereis dumerilii however, a Go-opsin is expressed in the rhabdomeric photoreceptor cells of the eyes.[87]
RGR-opsins
[edit]RGR-opsins, also known as Retinal G protein coupled receptors are expressed in the retinal pigment epithelium (RPE) and Müller cells.[104] They preferentially bind all-trans-retinal in the dark instead of 11-cis-retinal.[41] RGR-opsins were thought to be photoisomerases[44] but instead, they regulate retinoid traffic and production.[66][105] In particular, they speed up light-independently the production of 11-cis-retinol (a precursor of 11-cis-retinal) from all-trans-retinyl-esters.[106] However, the all-trans-retinyl-esters are made available light-dependently by RGR-opsins. Whether RGR-opsins regulate this via a G-protein or another signaling mechanism is unknown.[107] The cattle RGR-opsin absorbs maximally at different wavelengths depending on the pH-value. At high pH it absorbs maximally blue (469 nm) light and at low pH it absorbs maximally UV (370 nm) light.[108]
Peropsin
[edit]Peropsin, a visual pigment-like receptor, is a protein that in humans is encoded by the RRH gene.[109]
Other proteins called opsins
[edit]Photoreceptors can be classified several ways, including function (vision, phototaxis, photoperiodism, etc.), type of chromophore (retinal, flavine, bilin), molecular structure (tertiary, quaternary), signal output (phosphorylation, reduction, oxidation), etc.[110]
Beside animal opsins, which are G protein-coupled receptors, there is another group of photoreceptor proteins called opsins.[67][111] These are the microbial opsin, they are used by prokaryotes and by some algae (as a component of channelrhodopsins) and fungi,[112] whereas animals use animal opsins, exclusively. No opsins have been found outside these groups (for instance in plants, or placozoans).[67]
Microbial and animal opsins are also called type 1 and type 2 opsins respectively. Both types are called opsins, because at one time it was thought that they were related: Both are seven-transmembrane receptors and bind covalently retinal as chromophore, which turns them into photoreceptors sensing light. However, both types are not related on the sequence level.[113]
In fact, the sequence identity between animal and mirobial opsins is no greater than could be accounted for by random chance. However, in recent years new methods have been developed specific to deep phylogeny. As a result, several studies have found evidence of a possible phylogenetic relationship between the two.[114][35][115] However, this does not necessarily mean that the last common ancestor of microbial and animal opsins was itself light sensitive: All animal opsins arose (by gene duplication and divergence) late in the history of the large G-protein coupled receptor (GPCR) gene family, which itself arose after the divergence of plants, fungi, choanflagellates and sponges from the earliest animals. The retinal chromophore is found solely in the opsin branch of this large gene family, meaning its occurrence elsewhere represents convergent evolution, not homology. Microbial rhodopsins are, by sequence, very different from any of the GPCR families.[116] According to one hypothesis, both microbial and animal opsins belong to the transporter-opsin-G protein-coupled receptor (TOG) superfamily, a proposed clade that includes G protein-coupled receptor (GPCR), Ion-translocating microbial rhodopsin (MR), and seven others.[117]
Most microbial opsins are ion channels or pumps instead of proper receptors and do not bind to a G protein. Microbal opsins are found in all three domains of life: Archaea, Bacteria, and Eukaryota. In Eukaryota, microbial opsins are found mainly in unicellular organisms such as green algae, and in fungi. In most complex multicellular eukaryotes, microbial opsins have been replaced with other light-sensitive molecules such as cryptochrome and phytochrome in plants, and animal opsins in animals.[118]
Microbial opsins are often known by the rhodopsin form of the molecule, i.e., rhodopsin (in the broad sense) = opsin + chromophore. Among the many kinds of microbial opsins are the proton pumps bacteriorhodopsin (BR) and xanthorhodopsin (xR), the chloride pump halorhodopsin (HR), the photosensors sensory rhodopsin I (SRI) and sensory rhodopsin II (SRII), as well as proteorhodopsin (PR), Neurospora opsin I (NOPI), Chlamydomonas sensory rhodopsins A (CSRA), Chlamydomonas sensory rhodopsins B (CSRB), channelrhodopsin (ChR), and archaerhodopsin (Arch).[119]
Several microbal opsins, such as proteo- and bacteriorhodopsin, are used by various bacterial groups to harvest energy from light to carry out metabolic processes using a non-chlorophyll-based pathway. Beside that, halorhodopsins of Halobacteria and channelrhodopsins of some algae, e.g. Volvox, serve them as light-gated ion channels, amongst others also for phototactic purposes. Sensory rhodopsins exist in Halobacteria that induce a phototactic response by interacting with transducer membrane-embedded proteins that have no relation to G proteins.[120]
Microbal opsins (like channelrhodopsin, halorhodopsin, and archaerhodopsin) are used in optogenetics to switch on or off neuronal activity. Microbal opsins are preferred if the neuronal activity should be modulated at higher frequency, because they respond faster than animal opsins. This is because microbal opsins are ion channels or proton/ion pumps and thus are activated by light directly, while animal opsins activate G-proteins, which then activate effector enzymes that produce metabolites to open ion channels.[121]
See also
[edit]External links
[edit]- Illustration Archived 2020-01-09 at the Wayback Machine at Baldwin-Wallace College
- Opsin at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
References
[edit]- ^ a b Casey PJ, Gilman AG (February 1988). "G protein involvement in receptor-effector coupling". The Journal of Biological Chemistry. 263 (6): 2577–2580. doi:10.1016/s0021-9258(18)69103-3. PMID 2830256. S2CID 38970721.
- ^ a b Attwood TK, Findlay JB (February 1994). "Fingerprinting G-protein-coupled receptors". Protein Engineering. 7 (2): 195–203. doi:10.1093/protein/7.2.195. PMID 8170923.
- ^ Dixon RA, Kobilka BK, Strader DJ, Benovic JL, Dohlman HG, Frielle T, et al. (May 1986). "Cloning of the gene and cDNA for mammalian beta-adrenergic receptor and homology with rhodopsin". Nature. 321 (6065): 75–79. Bibcode:1986Natur.321...75D. doi:10.1038/321075a0. PMID 3010132. S2CID 4324074.
- ^ Dixon RA, Sigal IS, Rands E, Register RB, Candelore MR, Blake AD, Strader CD (March 1987). "Ligand binding to the beta-adrenergic receptor involves its rhodopsin-like core". Nature. 326 (6108): 73–77. Bibcode:1987Natur.326...73D. doi:10.1038/326073a0. PMID 2881211. S2CID 4352920.
- ^ Wald G (July 1934). "Carotenoids and the Vitamin A Cycle in Vision". Nature. 134 (3376): 65. Bibcode:1934Natur.134...65W. doi:10.1038/134065a0. S2CID 4022911.
- ^ Wald G, Brown PK, Hubbard R, Oroshnik W (July 1955). "Hindered Cis Isomers of Vitamin a and Retinene: The Structure of the Neo-B Isomer". Proceedings of the National Academy of Sciences of the United States of America. 41 (7): 438–451. Bibcode:1955PNAS...41..438W. doi:10.1073/pnas.41.7.438. PMC 528115. PMID 16589696.
- ^ Brown PK, Wald G (October 1956). "The neo-b isomer of vitamin A and retinene". The Journal of Biological Chemistry. 222 (2): 865–877. doi:10.1016/S0021-9258(20)89944-X. PMID 13367054.
- ^ Oroshnik W (June 1956). "The Synthesis and Configuration of Neo-B Vitamin A and Neoretinine b". Journal of the American Chemical Society. 78 (11): 2651–2652. doi:10.1021/ja01592a095.
- ^ Oroshnik W, Brown PK, Hubbard R, Wald G (September 1956). "HINDERED CIS ISOMERS OF VITAMIN A AND RETINENE: THE STRUCTURE OF THE NEO-b ISOMER". Proceedings of the National Academy of Sciences of the United States of America. 42 (9): 578–580. Bibcode:1956PNAS...42..578O. doi:10.1073/pnas.42.9.578. PMC 534254. PMID 16589909.
- ^ Bownds D (December 1967). "Site of attachment of retinal in rhodopsin". Nature. 216 (5121): 1178–1181. Bibcode:1967Natur.216.1178B. doi:10.1038/2161178a0. PMID 4294735. S2CID 1657759.
- ^ Hargrave PA, McDowell JH, Curtis DR, Wang JK, Juszczak E, Fong SL, et al. (1983). "The structure of bovine rhodopsin". Biophysics of Structure and Mechanism. 9 (4): 235–244. doi:10.1007/BF00535659. PMID 6342691. S2CID 20407577.
- ^ a b c Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, et al. (August 2000). "Crystal structure of rhodopsin: A G protein-coupled receptor". Science. 289 (5480): 739–745. Bibcode:2000Sci...289..739P. CiteSeerX 10.1.1.1012.2275. doi:10.1126/science.289.5480.739. PMID 10926528.
- ^ a b Murakami M, Kouyama T (May 2008). "Crystal structure of squid rhodopsin". Nature. 453 (7193): 363–367. Bibcode:2008Natur.453..363M. doi:10.1038/nature06925. PMID 18480818. S2CID 4339970.
- ^ Collins FD (March 1953). "Rhodopsin and indicator yellow". Nature. 171 (4350): 469–471. Bibcode:1953Natur.171..469C. doi:10.1038/171469a0. PMID 13046517. S2CID 4152360.
- ^ Pitt GA, Collins FD, Morton RA, Stok P (January 1955). "Studies on rhodopsin. VIII. Retinylidenemethylamine, an indicator yellow analogue". The Biochemical Journal. 59 (1): 122–128. doi:10.1042/bj0590122. PMC 1216098. PMID 14351151.
- ^ Hubbard R, Kropf A (February 1958). "The Action of Light on Rhodopsin". Proceedings of the National Academy of Sciences of the United States of America. 44 (2): 130–139. Bibcode:1958PNAS...44..130H. doi:10.1073/pnas.44.2.130. PMC 335377. PMID 16590155.
- ^ Kropf A, Hubbard R (November 1959). "The mechanism of bleaching rhodopsin". Annals of the New York Academy of Sciences. 74 (2): 266–280. Bibcode:1959NYASA..74..266K. doi:10.1111/j.1749-6632.1958.tb39550.x. PMID 13627857. S2CID 45830716.
- ^ a b Choe HW, Kim YJ, Park JH, Morizumi T, Pai EF, Krauss N, et al. (March 2011). "Crystal structure of metarhodopsin II". Nature. 471 (7340): 651–655. Bibcode:2011Natur.471..651C. doi:10.1038/nature09789. PMID 21389988. S2CID 4302421.
- ^ a b Wald G (October 1968). "Molecular basis of visual excitation". Science. 162 (3850): 230–239. Bibcode:1968Sci...162..230W. doi:10.1126/science.162.3850.230. PMID 4877437.
- ^ Terakita A, Kawano-Yamashita E, Koyanagi M (January 2012). "Evolution and diversity of opsins". Wiley Interdisciplinary Reviews: Membrane Transport and Signaling. 1 (1): 104–111. doi:10.1002/wmts.6.
- ^ a b c d e f g h i j k l m n o p q r s t u Gühmann M, Porter ML, Bok MJ (August 2022). "The Gluopsins: Opsins without the Retinal Binding Lysine". Cells. 11 (15): 2441. doi:10.3390/cells11152441. PMC 9368030. PMID 35954284.
 Material was copied and adapted from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied and adapted from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ Amora TL, Ramos LS, Galan JF, Birge RR (April 2008). "Spectral tuning of deep red cone pigments". Biochemistry. 47 (16): 4614–4620. doi:10.1021/bi702069d. PMC 2492582. PMID 18370404.
- ^ Ballesteros JA, Weinstein H (1995). "Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors". Methods in Neurosciences. 25: 366–428. doi:10.1016/S1043-9471(05)80049-7. ISBN 978-0-12-185295-5.
- ^ a b Ovchinnikov, Yu.A. (November 1982). "Rhodopsin and bacteriorhodopsin: structure-function relationships". FEBS Letters. 148 (2): 179–191. doi:10.1016/0014-5793(82)80805-3. PMID 6759163. S2CID 85819100.
- ^ a b Katana R, Guan C, Zanini D, Larsen ME, Giraldo D, Geurten BR, et al. (September 2019). "Chromophore-Independent Roles of Opsin Apoproteins in Drosophila Mechanoreceptors". Current Biology. 29 (17): 2961–2969.e4. Bibcode:2019CBio...29E2961K. doi:10.1016/j.cub.2019.07.036. PMID 31447373. S2CID 201420079.
- ^ a b Leung NY, Thakur DP, Gurav AS, Kim SH, Di Pizio A, Niv MY, Montell C (April 2020). "Functions of Opsins in Drosophila Taste". Current Biology. 30 (8): 1367–1379.e6. Bibcode:2020CBio...30E1367L. doi:10.1016/j.cub.2020.01.068. PMC 7252503. PMID 32243853.
- ^ a b c d e Porter ML, Blasic JR, Bok MJ, Cameron EG, Pringle T, Cronin TW, Robinson PR (January 2012). "Shedding new light on opsin evolution". Proceedings. Biological Sciences. 279 (1726): 3–14. doi:10.1098/rspb.2011.1819. PMC 3223661. PMID 22012981.
- ^ a b c d e f g h Ramirez MD, Pairett AN, Pankey MS, Serb JM, Speiser DI, Swafford AJ, Oakley TH (December 2016). "The Last Common Ancestor of Most Bilaterian Animals Possessed at Least Nine Opsins". Genome Biology and Evolution. 8 (12): 3640–3652. doi:10.1093/gbe/evw248. PMC 5521729. PMID 28172965.
- ^ a b Troemel ER, Chou JH, Dwyer ND, Colbert HA, Bargmann CI (October 1995). "Divergent seven transmembrane receptors are candidate chemosensory receptors in C. elegans". Cell. 83 (2): 207–218. doi:10.1016/0092-8674(95)90162-0. PMID 7585938. S2CID 17819587.
- ^ D'Aniello S, Delroisse J, Valero-Gracia A, Lowe EK, Byrne M, Cannon JT, et al. (December 2015). "Opsin evolution in the Ambulacraria". Marine Genomics. 24 (Pt 2): 177–183. Bibcode:2015MarGn..24..177D. doi:10.1016/j.margen.2015.10.001. PMID 26472700.
- ^ Shen WL, Kwon Y, Adegbola AA, Luo J, Chess A, Montell C (March 2011). "Function of rhodopsin in temperature discrimination in Drosophila". Science. 331 (6022): 1333–1336. Bibcode:2011Sci...331.1333S. doi:10.1126/science.1198904. PMID 21393546. S2CID 206530389.
- ^ Senthilan PR, Piepenbrock D, Ovezmyradov G, Nadrowski B, Bechstedt S, Pauls S, et al. (August 2012). "Drosophila auditory organ genes and genetic hearing defects". Cell. 150 (5): 1042–1054. doi:10.1016/j.cell.2012.06.043. PMID 22939627. S2CID 1422764.
- ^ Feuda R, Menon AK, Göpfert MC (March 2022). "Rethinking Opsins". Molecular Biology and Evolution. 39 (3): msac033. doi:10.1093/molbev/msac033. PMC 8892948. PMID 35143663.
- ^ a b Leung NY, Montell C (October 2017). "Unconventional Roles of Opsins". Annual Review of Cell and Developmental Biology. 33 (1): 241–264. doi:10.1146/annurev-cellbio-100616-060432. PMC 5963513. PMID 28598695.
- ^ a b Devine EL, Oprian DD, Theobald DL (August 2013). "Relocating the active-site lysine in rhodopsin and implications for evolution of retinylidene proteins". Proceedings of the National Academy of Sciences of the United States of America. 110 (33): 13351–13355. Bibcode:2013PNAS..11013351D. doi:10.1073/pnas.1306826110. PMC 3746867. PMID 23904486.
- ^ Borroto-Escuela DO, Romero-Fernandez W, García-Negredo G, Correia PA, Garriga P, Fuxe K, Ciruela F (2011). "Dissecting the conserved NPxxY motif of the M3 muscarinic acetylcholine receptor: critical role of Asp-7.49 for receptor signaling and multiprotein complex formation". Cellular Physiology and Biochemistry. 28 (5): 1009–1022. doi:10.1159/000335788. hdl:2445/126278. PMID 22178951. S2CID 14008354.
- ^ a b Fritze O, Filipek S, Kuksa V, Palczewski K, Hofmann KP, Ernst OP (March 2003). "Role of the conserved NPxxY(x)5,6F motif in the rhodopsin ground state and during activation". Proceedings of the National Academy of Sciences of the United States of America. 100 (5): 2290–2295. Bibcode:2003PNAS..100.2290F. doi:10.1073/pnas.0435715100. PMC 151333. PMID 12601165.
- ^ Zhou Q, Yang D, Wu M, Guo Y, Guo W, Zhong L, et al. (December 2019). "Common activation mechanism of class A GPCRs". eLife. 8: e50279. doi:10.7554/eLife.50279. PMC 6954041. PMID 31855179.
- ^ Wess J, Nanavati S, Vogel Z, Maggio R (January 1993). "Functional role of proline and tryptophan residues highly conserved among G protein-coupled receptors studied by mutational analysis of the m3 muscarinic receptor". The EMBO Journal. 12 (1): 331–338. doi:10.1002/j.1460-2075.1993.tb05661.x. PMC 413210. PMID 7679072.
- ^ Galés C, Kowalski-Chauvel A, Dufour MN, Seva C, Moroder L, Pradayrol L, et al. (June 2000). "Mutation of Asn-391 within the conserved NPXXY motif of the cholecystokinin B receptor abolishes Gq protein activation without affecting its association with the receptor". The Journal of Biological Chemistry. 275 (23): 17321–17327. doi:10.1074/jbc.M909801199. PMID 10748160.
- ^ a b Hao W, Fong HK (March 1999). "The endogenous chromophore of retinal G protein-coupled receptor opsin from the pigment epithelium". The Journal of Biological Chemistry. 274 (10): 6085–6090. doi:10.1074/jbc.274.10.6085. PMID 10037690.
- ^ Hara T, Hara R (May 1967). "Rhodopsin and retinochrome in the squid retina". Nature. 214 (5088): 573–575. Bibcode:1967Natur.214..573H. doi:10.1038/214573a0. PMID 6036171. S2CID 4184319.
- ^ a b Tsukamoto H, Terakita A (November 2010). "Diversity and functional properties of bistable pigments". Photochemical & Photobiological Sciences. 9 (11): 1435–1443. doi:10.1039/c0pp00168f. PMID 20852774.
- ^ a b c d e f g h i j k l Terakita A (1 March 2005). "The opsins". Genome Biology. 6 (3): 213. doi:10.1186/gb-2005-6-3-213. PMC 1088937. PMID 15774036.
- ^ Nagata T, Koyanagi M, Tsukamoto H, Terakita A (January 2010). "Identification and characterization of a protostome homologue of peropsin from a jumping spider". Journal of Comparative Physiology A. 196 (1): 51–59. doi:10.1007/s00359-009-0493-9. PMID 19960196. S2CID 22879394.
- ^ Gehring WJ (January 2014). "The evolution of vision". Wiley Interdisciplinary Reviews. Developmental Biology. 3 (1): 1–40. doi:10.1002/wdev.96. PMID 24902832. S2CID 36881435.
- ^ a b c Kato M, Sugiyama T, Sakai K, Yamashita T, Fujita H, Sato K, et al. (18 November 2016). "Two Opsin 3-Related Proteins in the Chicken Retina and Brain: A TMT-Type Opsin 3 Is a Blue-Light Sensor in Retinal Horizontal Cells, Hypothalamus, and Cerebellum". PLOS ONE. 11 (11): e0163925. Bibcode:2016PLoSO..1163925K. doi:10.1371/journal.pone.0163925. PMC 5115664. PMID 27861495.
- ^ Mazna P, Grycova L, Balik A, Zemkova H, Friedlova E, Obsilova V, et al. (November 2008). "The role of proline residues in the structure and function of human MT2 melatonin receptor". Journal of Pineal Research. 45 (4): 361–372. doi:10.1111/j.1600-079X.2008.00598.x. PMID 18544139. S2CID 6202186.
- ^ Yokoyama S (July 2000). "Molecular evolution of vertebrate visual pigments". Progress in Retinal and Eye Research. 19 (4): 385–419. doi:10.1016/S1350-9462(00)00002-1. PMID 10785616. S2CID 28746630.
- ^ Deeb SS (May 2005). "The molecular basis of variation in human color vision". Clinical Genetics. 67 (5): 369–377. doi:10.1111/j.1399-0004.2004.00343.x. PMID 15811001. S2CID 24105079.
- ^ a b c d e f Haltaufderhyde K, Ozdeslik RN, Wicks NL, Najera JA, Oancea E (2015). "Opsin expression in human epidermal skin". Photochemistry and Photobiology. 91 (1): 117–123. doi:10.1111/php.12354. PMC 4303996. PMID 25267311.
- ^ White JH, Chiano M, Wigglesworth M, Geske R, Riley J, White N, et al. (July 2008). "Identification of a novel asthma susceptibility gene on chromosome 1qter and its functional evaluation". Human Molecular Genetics. 17 (13): 1890–1903. doi:10.1093/hmg/ddn087. PMID 18344558.
- ^ a b c d Blackshaw S, Snyder SH (May 1999). "Encephalopsin: a novel mammalian extraretinal opsin discretely localized in the brain". The Journal of Neuroscience. 19 (10): 3681–3690. doi:10.1523/JNEUROSCI.19-10-03681.1999. PMC 6782724. PMID 10234000.
- ^ Nissilä J, Mänttäri S, Särkioja T, Tuominen H, Takala T, Timonen M, Saarela S (November 2012). "Encephalopsin (OPN3) protein abundance in the adult mouse brain". Journal of Comparative Physiology A. 198 (11): 833–839. doi:10.1007/s00359-012-0754-x. PMC 3478508. PMID 22991144.
- ^ a b Bailes HJ, Lucas RJ (May 2013). "Human melanopsin forms a pigment maximally sensitive to blue light (λmax ≈ 479 nm) supporting activation of G(q/11) and G(i/o) signalling cascades". Proceedings. Biological Sciences. 280 (1759): 20122987. doi:10.1098/rspb.2012.2987. PMC 3619500. PMID 23554393.
- ^ a b c d Kojima D, Mori S, Torii M, Wada A, Morishita R, Fukada Y (17 October 2011). "UV-sensitive photoreceptor protein OPN5 in humans and mice". PLOS ONE. 6 (10): e26388. Bibcode:2011PLoSO...626388K. doi:10.1371/journal.pone.0026388. PMC 3197025. PMID 22043319.
- ^ Tarttelin EE, Bellingham J, Hankins MW, Foster RG, Lucas RJ (November 2003). "Neuropsin (Opn5): a novel opsin identified in mammalian neural tissue". FEBS Letters. 554 (3): 410–416. doi:10.1016/S0014-5793(03)01212-2. PMID 14623103.
- ^ Yamashita T, Ono K, Ohuchi H, Yumoto A, Gotoh H, Tomonari S, et al. (February 2014). "Evolution of mammalian Opn5 as a specialized UV-absorbing pigment by a single amino acid mutation". The Journal of Biological Chemistry. 289 (7): 3991–4000. doi:10.1074/jbc.M113.514075. PMC 3924266. PMID 24403072.
- ^ Mäthger LM, Roberts SB, Hanlon RT (October 2010). "Evidence for distributed light sensing in the skin of cuttlefish, Sepia officinalis". Biology Letters. 6 (5): 600–603. doi:10.1098/rsbl.2010.0223. PMC 2936158. PMID 20392722.
- ^ Yong E (20 May 2015). "Octopuses, and Maybe Squid, Can Sense Light With Their Skin". National Geographic. Archived from the original on February 23, 2021.
- ^ Yu C, Li Y, Zhang X, Huang X, Malyarchuk V, Wang S, et al. (September 2014). "Adaptive optoelectronic camouflage systems with designs inspired by cephalopod skins". Proceedings of the National Academy of Sciences of the United States of America. 111 (36): 12998–13003. Bibcode:2014PNAS..11112998Y. doi:10.1073/pnas.1410494111. PMC 4246966. PMID 25136094.
- ^ a b Liegertová M, Pergner J, Kozmiková I, Fabian P, Pombinho AR, Strnad H, et al. (July 2015). "Cubozoan genome illuminates functional diversification of opsins and photoreceptor evolution". Scientific Reports. 5: 11885. Bibcode:2015NatSR...511885L. doi:10.1038/srep11885. PMC 5155618. PMID 26154478.
- ^ Quiroga Artigas G, Lapébie P, Leclère L, Takeda N, Deguchi R, Jékely G, et al. (January 2018). "A gonad-expressed opsin mediates light-induced spawning in the jellyfish Clytia". eLife. 7: e29555. doi:10.7554/eLife.29555. PMC 5756024. PMID 29303477.
- ^ a b c Rawlinson KA, Lapraz F, Ballister ER, Terasaki M, Rodgers J, McDowell RJ, et al. (October 2019). "Extraocular, rod-like photoreceptors in a flatworm express xenopsin photopigment". eLife. 8: e45465. doi:10.7554/eLife.45465. PMC 6805122. PMID 31635694.
- ^ a b Feuda R, Hamilton SC, McInerney JO, Pisani D (November 2012). "Metazoan opsin evolution reveals a simple route to animal vision". Proceedings of the National Academy of Sciences of the United States of America. 109 (46): 18868–18872. Bibcode:2012PNAS..10918868F. doi:10.1073/pnas.1204609109. PMC 3503164. PMID 23112152.
- ^ a b c d e f Shichida Y, Matsuyama T (October 2009). "Evolution of opsins and phototransduction". Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 364 (1531): 2881–2895. doi:10.1098/rstb.2009.0051. PMC 2781858. PMID 19720651.
- ^ a b c d Plachetzki DC, Fong CR, Oakley TH (July 2010). "The evolution of phototransduction from an ancestral cyclic nucleotide gated pathway". Proceedings. Biological Sciences. 277 (1690): 1963–1969. doi:10.1098/rspb.2009.1797. PMC 2880087. PMID 20219739.
- ^ Hunt DM, Carvalho LS, Cowing JA, Davies WL (October 2009). "Evolution and spectral tuning of visual pigments in birds and mammals". Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 364 (1531): 2941–2955. doi:10.1098/rstb.2009.0044. PMC 2781856. PMID 19720655.
- ^ Trezise AE, Collin SP (October 2005). "Opsins: evolution in waiting". Current Biology. 15 (19): R794–R796. Bibcode:2005CBio...15.R794T. doi:10.1016/j.cub.2005.09.025. PMID 16213808.
- ^ Gulati S, Jastrzebska B, Banerjee S, Placeres ÁL, Miszta P, Gao S, et al. (March 2017). "Photocyclic behavior of rhodopsin induced by an atypical isomerization mechanism". Proceedings of the National Academy of Sciences of the United States of America. 114 (13): E2608–E2615. Bibcode:2017PNAS..114E2608G. doi:10.1073/pnas.1617446114. PMC 5380078. PMID 28289214.
- ^ Mano H, Kojima D, Fukada Y (November 1999). "Exo-rhodopsin: a novel rhodopsin expressed in the zebrafish pineal gland". Brain Research. Molecular Brain Research. 73 (1–2): 110–118. doi:10.1016/S0169-328X(99)00242-9. PMID 10581404.
- ^ Tarttelin EE, Fransen MP, Edwards PC, Hankins MW, Schertler GF, Vogel R, et al. (November 2011). "Adaptation of pineal expressed teleost exo-rod opsin to non-image forming photoreception through enhanced Meta II decay". Cellular and Molecular Life Sciences. 68 (22): 3713–3723. doi:10.1007/s00018-011-0665-y. PMC 3203999. PMID 21416149.
- ^ Okano T, Yoshizawa T, Fukada Y (November 1994). "Pinopsin is a chicken pineal photoreceptive molecule". Nature. 372 (6501): 94–97. Bibcode:1994Natur.372...94O. doi:10.1038/372094a0. PMID 7969427. S2CID 4301315.
- ^ Nakane Y, Yoshimura T (February 2019). "Photoperiodic Regulation of Reproduction in Vertebrates". Annual Review of Animal Biosciences. 7 (1). Annual Reviews: 173–194. doi:10.1146/annurev-animal-020518-115216. PMID 30332291. S2CID 52984435.
- ^ Philp AR, Garcia-Fernandez JM, Soni BG, Lucas RJ, Bellingham J, Foster RG (June 2000). "Vertebrate ancient (VA) opsin and extraretinal photoreception in the Atlantic salmon (Salmo salar)". The Journal of Experimental Biology. 203 (Pt 12): 1925–1936. doi:10.1242/jeb.203.12.1925. PMID 10821749.
- ^ Poletini MO, Ramos BC, Moraes MN, Castrucci AM (2015). "Nonvisual Opsins and the Regulation of Peripheral Clocks by Light and Hormones". Photochemistry and Photobiology. 91 (5): 1046–1055. doi:10.1111/php.12494. PMID 26174318. S2CID 41895317.
- ^ Blackshaw S, Snyder SH (November 1997). "Parapinopsin, a novel catfish opsin localized to the parapineal organ, defines a new gene family". The Journal of Neuroscience. 17 (21): 8083–8092. doi:10.1523/JNEUROSCI.17-21-08083.1997. PMC 6573767. PMID 9334384.
- ^ Koyanagi M, Kawano E, Kinugawa Y, Oishi T, Shichida Y, Tamotsu S, Terakita A (April 2004). "Bistable UV pigment in the lamprey pineal". Proceedings of the National Academy of Sciences of the United States of America. 101 (17): 6687–6691. Bibcode:2004PNAS..101.6687K. doi:10.1073/pnas.0400819101. PMC 404106. PMID 15096614.
- ^ Koyanagi M, Wada S, Kawano-Yamashita E, Hara Y, Kuraku S, Kosaka S, et al. (September 2015). "Diversification of non-visual photopigment parapinopsin in spectral sensitivity for diverse pineal functions". BMC Biology. 13 (1): 73. doi:10.1186/s12915-015-0174-9. PMC 4570685. PMID 26370232.
- ^ Su CY, Luo DG, Terakita A, Shichida Y, Liao HW, Kazmi MA, et al. (March 2006). "Parietal-eye phototransduction components and their potential evolutionary implications". Science. 311 (5767): 1617–1621. Bibcode:2006Sci...311.1617S. doi:10.1126/science.1123802. PMID 16543463. S2CID 28604455.
- ^ Koyanagi M, Terakita A (May 2014). "Diversity of animal opsin-based pigments and their optogenetic potential". Biochimica et Biophysica Acta (BBA) - Bioenergetics. 1837 (5): 710–716. doi:10.1016/j.bbabio.2013.09.003. PMID 24041647.
- ^ a b c Halford S, Freedman MS, Bellingham J, Inglis SL, Poopalasundaram S, Soni BG, et al. (March 2001). "Characterization of a novel human opsin gene with wide tissue expression and identification of embedded and flanking genes on chromosome 1q43". Genomics. 72 (2): 203–208. doi:10.1006/geno.2001.6469. PMID 11401433.
- ^ Arendt D, Tessmar-Raible K, Snyman H, Dorresteijn AW, Wittbrodt J (October 2004). "Ciliary photoreceptors with a vertebrate-type opsin in an invertebrate brain". Science. 306 (5697): 869–871. Bibcode:2004Sci...306..869A. doi:10.1126/science.1099955. PMID 15514158. S2CID 2583520.
- ^ Tsukamoto H, Chen IS, Kubo Y, Furutani Y (August 2017). "A ciliary opsin in the brain of a marine annelid zooplankton is ultraviolet-sensitive, and the sensitivity is tuned by a single amino acid residue". The Journal of Biological Chemistry. 292 (31): 12971–12980. doi:10.1074/jbc.M117.793539. PMC 5546036. PMID 28623234.
- ^ Ayers T, Tsukamoto H, Gühmann M, Veedin Rajan VB, Tessmar-Raible K (April 2018). "A Go-type opsin mediates the shadow reflex in the annelid Platynereis dumerilii". BMC Biology. 16 (1): 41. doi:10.1186/s12915-018-0505-8. PMC 5904973. PMID 29669554.
- ^ Verasztó C, Gühmann M, Jia H, Rajan VB, Bezares-Calderón LA, Piñeiro-Lopez C, et al. (May 2018). "Ciliary and rhabdomeric photoreceptor-cell circuits form a spectral depth gauge in marine zooplankton". eLife. 7. doi:10.7554/eLife.36440. PMC 6019069. PMID 29809157.
- ^ a b Gühmann M, Jia H, Randel N, Verasztó C, Bezares-Calderón LA, Michiels NK, et al. (August 2015). "Spectral Tuning of Phototaxis by a Go-Opsin in the Rhabdomeric Eyes of Platynereis". Current Biology. 25 (17): 2265–2271. Bibcode:2015CBio...25.2265G. doi:10.1016/j.cub.2015.07.017. PMID 26255845.
- ^ Nilsson DE (October 2009). "The evolution of eyes and visually guided behaviour". Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 364 (1531): 2833–2847. doi:10.1098/rstb.2009.0083. PMC 2781862. PMID 19720648.
- ^ Nilsson DE (March 2013). "Eye evolution and its functional basis". Visual Neuroscience. 30 (1–2): 5–20. doi:10.1017/S0952523813000035. PMC 3632888. PMID 23578808.
- ^ Koyanagi M, Takada E, Nagata T, Tsukamoto H, Terakita A (March 2013). "Homologs of vertebrate Opn3 potentially serve as a light sensor in nonphotoreceptive tissue". Proceedings of the National Academy of Sciences of the United States of America. 110 (13): 4998–5003. Bibcode:2013PNAS..110.4998K. doi:10.1073/pnas.1219416110. PMC 3612648. PMID 23479626.
- ^ a b Sakai K, Yamashita T, Imamoto Y, Shichida Y (22 October 2015). "Diversity of Active States in TMT Opsins". PLOS ONE. 10 (10): e0141238. Bibcode:2015PLoSO..1041238S. doi:10.1371/journal.pone.0141238. PMC 4619619. PMID 26491964.
- ^ a b Fischer RM, Fontinha BM, Kirchmaier S, Steger J, Bloch S, Inoue D, et al. (11 June 2013). "Co-expression of VAL- and TMT-opsins uncovers ancient photosensory interneurons and motorneurons in the vertebrate brain". PLOS Biology. 11 (6): e1001585. doi:10.1371/journal.pbio.1001585. PMC 3679003. PMID 23776409.
- ^ a b Moutsaki P, Whitmore D, Bellingham J, Sakamoto K, David-Gray ZK, Foster RG (April 2003). "Teleost multiple tissue (tmt) opsin: a candidate photopigment regulating the peripheral clocks of zebrafish?". Brain Research. Molecular Brain Research. 112 (1–2): 135–145. doi:10.1016/S0169-328X(03)00059-7. PMID 12670711.
- ^ Koyanagi M, Takano K, Tsukamoto H, Ohtsu K, Tokunaga F, Terakita A (October 2008). "Jellyfish vision starts with cAMP signaling mediated by opsin-G(s) cascade". Proceedings of the National Academy of Sciences of the United States of America. 105 (40): 15576–15580. Bibcode:2008PNAS..10515576K. doi:10.1073/pnas.0806215105. PMC 2563118. PMID 18832159.
- ^ Mason B, Schmale M, Gibbs P, Miller MW, Wang Q, Levay K, et al. (5 December 2012). "Evidence for multiple phototransduction pathways in a reef-building coral". PLOS ONE. 7 (12): e50371. Bibcode:2012PLoSO...750371M. doi:10.1371/journal.pone.0050371. PMC 3515558. PMID 23227169.
- ^ Suga H, Schmid V, Gehring WJ (January 2008). "Evolution and functional diversity of jellyfish opsins". Current Biology. 18 (1): 51–55. Bibcode:2008CBio...18...51S. doi:10.1016/j.cub.2007.11.059. PMID 18160295.
- ^ Feuda R, Rota-Stabelli O, Oakley TH, Pisani D (July 2014). "The comb jelly opsins and the origins of animal phototransduction". Genome Biology and Evolution. 6 (8): 1964–1971. doi:10.1093/gbe/evu154. PMC 4159004. PMID 25062921.
- ^ Smith WC, Price DA, Greenberg RM, Battelle BA (July 1993). "Opsins from the lateral eyes and ocelli of the horseshoe crab, Limulus polyphemus". Proceedings of the National Academy of Sciences of the United States of America. 90 (13): 6150–6154. Bibcode:1993PNAS...90.6150S. doi:10.1073/pnas.90.13.6150. PMC 46885. PMID 8327495.
- ^ a b Yamashita T, Ohuchi H, Tomonari S, Ikeda K, Sakai K, Shichida Y (December 2010). "Opn5 is a UV-sensitive bistable pigment that couples with Gi and Gq subtype of G protein". Proceedings of the National Academy of Sciences of the United States of America. 107 (51): 22084–22089. Bibcode:2010PNAS..10722084Y. doi:10.1073/pnas.1012498107. PMC 3009823. PMID 21135214.
- ^ Buhr ED, Yue WW, Ren X, Jiang Z, Liao HW, Mei X, et al. (October 2015). "Neuropsin (OPN5)-mediated photoentrainment of local circadian oscillators in mammalian retina and cornea". Proceedings of the National Academy of Sciences of the United States of America. 112 (42): 13093–13098. Bibcode:2015PNAS..11213093B. doi:10.1073/pnas.1516259112. PMC 4620855. PMID 26392540.
- ^ Hering L, Mayer G (September 2014). "Analysis of the opsin repertoire in the tardigrade Hypsibius dujardini provides insights into the evolution of opsin genes in panarthropoda". Genome Biology and Evolution. 6 (9): 2380–2391. doi:10.1093/gbe/evu193. PMC 4202329. PMID 25193307.
- ^ Kojima D, Terakita A, Ishikawa T, Tsukahara Y, Maeda A, Shichida Y (September 1997). "A novel Go-mediated phototransduction cascade in scallop visual cells". The Journal of Biological Chemistry. 272 (37): 22979–22982. doi:10.1074/jbc.272.37.22979. PMID 9287291.
- ^ Koyanagi M, Terakita A, Kubokawa K, Shichida Y (November 2002). "Amphioxus homologs of Go-coupled rhodopsin and peropsin having 11-cis- and all-trans-retinals as their chromophores". FEBS Letters. 531 (3): 525–528. doi:10.1016/s0014-5793(02)03616-5. PMID 12435605. S2CID 11669142.
- ^ Jiang M, Pandey S, Fong HK (December 1993). "An opsin homologue in the retina and pigment epithelium". Investigative Ophthalmology & Visual Science. 34 (13): 3669–3678. PMID 8258527.
- ^ Nagata T, Koyanagi M, Terakita A (20 October 2010). "Molecular Evolution and Functional Diversity of Opsin-Based Photopigments". Retrieved 7 May 2018.
- ^ Wenzel A, Oberhauser V, Pugh EN, Lamb TD, Grimm C, Samardzija M, et al. (August 2005). "The retinal G protein-coupled receptor (RGR) enhances isomerohydrolase activity independent of light". The Journal of Biological Chemistry. 280 (33): 29874–29884. doi:10.1074/jbc.M503603200. PMID 15961402.
- ^ Radu RA, Hu J, Peng J, Bok D, Mata NL, Travis GH (July 2008). "Retinal pigment epithelium-retinal G protein receptor-opsin mediates light-dependent translocation of all-trans-retinyl esters for synthesis of visual chromophore in retinal pigment epithelial cells". The Journal of Biological Chemistry. 283 (28): 19730–19738. doi:10.1074/jbc.M801288200. PMC 2443657. PMID 18474598.
- ^ Hao W, Fong HK (May 1996). "Blue and ultraviolet light-absorbing opsin from the retinal pigment epithelium". Biochemistry. 35 (20): 6251–6256. doi:10.1021/bi952420k. PMID 8639565.
- ^ Sun H, Gilbert DJ, Copeland NG, Jenkins NA, Nathans J (September 1997). "Peropsin, a novel visual pigment-like protein located in the apical microvilli of the retinal pigment epithelium". Proceedings of the National Academy of Sciences of the United States of America. 94 (18): 9893–9898. Bibcode:1997PNAS...94.9893S. doi:10.1073/pnas.94.18.9893. PMC 23288. PMID 9275222.
- ^ Björn LO (2 January 2015). Photobiology: The Science of Light and Life. Springer. p. 169. ISBN 978-1-4939-1468-5. Retrieved 3 September 2015.
- ^ Fernald RD (September 2006). "Casting a genetic light on the evolution of eyes". Science. 313 (5795): 1914–1918. Bibcode:2006Sci...313.1914F. doi:10.1126/science.1127889. PMID 17008522. S2CID 84439732.
- ^ Waschuk SA, Bezerra AG, Shi L, Brown LS (May 2005). "Leptosphaeria rhodopsin: bacteriorhodopsin-like proton pump from a eukaryote". Proceedings of the National Academy of Sciences of the United States of America. 102 (19): 6879–6883. Bibcode:2005PNAS..102.6879W. doi:10.1073/pnas.0409659102. PMC 1100770. PMID 15860584.
- ^ Findlay JB, Pappin DJ (September 1986). "The opsin family of proteins". The Biochemical Journal. 238 (3): 625–642. doi:10.1042/bj2380625. PMC 1147185. PMID 2948499.
- ^ Shen L, Chen C, Zheng H, Jin L (2013). "The evolutionary relationship between microbial rhodopsins and metazoan rhodopsins". TheScientificWorldJournal. 2013: 435651. doi:10.1155/2013/435651. PMC 3583139. PMID 23476135.
- ^ Zhang Z, Jin Z, Zhao Y, Zhang Z, Li R, Xiao J, Wu J (August 2014). "Systematic study on G-protein couple receptor prototypes: did they really evolve from prokaryotic genes?". IET Systems Biology. 8 (4): 154–161. doi:10.1049/iet-syb.2013.0037. PMC 8687355. PMID 25075528.
- ^ Nordström KJ, Sällman Almén M, Edstam MM, Fredriksson R, Schiöth HB (September 2011). "Independent HHsearch, Needleman--Wunsch-based, and motif analyses reveal the overall hierarchy for most of the G protein-coupled receptor families". Molecular Biology and Evolution. 28 (9): 2471–2480. doi:10.1093/molbev/msr061. PMID 21402729.
- ^ Yee DC, Shlykov MA, Västermark A, Reddy VS, Arora S, Sun EI, Saier MH (November 2013). "The transporter-opsin-G protein-coupled receptor (TOG) superfamily". The FEBS Journal. 280 (22): 5780–5800. doi:10.1111/febs.12499. PMC 3832197. PMID 23981446.
- ^ Yoshizawa S, Kumagai Y, Kim H, Ogura Y, Hayashi T, Iwasaki W, et al. (May 2014). "Functional characterization of flavobacteria rhodopsins reveals a unique class of light-driven chloride pump in bacteria". Proceedings of the National Academy of Sciences of the United States of America. 111 (18): 6732–6737. Bibcode:2014PNAS..111.6732Y. doi:10.1073/pnas.1403051111. PMC 4020065. PMID 24706784.
- ^ Grote M, Engelhard M, Hegemann P (May 2014). "Of ion pumps, sensors and channels - perspectives on microbial rhodopsins between science and history". Biochimica et Biophysica Acta (BBA) - Bioenergetics. 1837 (5): 533–545. doi:10.1016/j.bbabio.2013.08.006. PMID 23994288.
- ^ Römpler H, Stäubert C, Thor D, Schulz A, Hofreiter M, Schöneberg T (February 2007). "G protein-coupled time travel: evolutionary aspects of GPCR research". Molecular Interventions. 7 (1): 17–25. doi:10.1124/mi.7.1.5. PMID 17339603.
- ^ Zhang F, Vierock J, Yizhar O, Fenno LE, Tsunoda S, Kianianmomeni A, et al. (December 2011). "The microbial opsin family of optogenetic tools". Cell. 147 (7): 1446–1457. doi:10.1016/j.cell.2011.12.004. PMC 4166436. PMID 22196724.



