G protein-coupled receptor: Difference between revisions
m Journal cites, added 2 PMCs, added 3 issue numbers, using AWB (8069) |
→Receptor structure: mentioned there are other 7tm receptors that are not G-protein coupled (see talk) |
||
| Line 58: | Line 58: | ||
GPCRs are [[integral membrane protein]]s that possess seven membrane-spanning domains or [[transmembrane helix|transmembrane helices]]. The extracellular parts of the receptor can be [[Glycosylation|glycosylated]]. These extracellular loops also contain two highly-conserved [[cysteine]] residues that form [[disulfide bond]]s to stabilize the receptor structure. Some seven-transmembrane helix proteins ([[channelrhodopsin]]) that resemble GPCRs may contain ion channels, within their protein. |
GPCRs are [[integral membrane protein]]s that possess seven membrane-spanning domains or [[transmembrane helix|transmembrane helices]]. The extracellular parts of the receptor can be [[Glycosylation|glycosylated]]. These extracellular loops also contain two highly-conserved [[cysteine]] residues that form [[disulfide bond]]s to stabilize the receptor structure. Some seven-transmembrane helix proteins ([[channelrhodopsin]]) that resemble GPCRs may contain ion channels, within their protein. |
||
Similar to (GPCRs, the adiponectin receptors 1 and 2 ([[ADIPOR1]] and [[ADIPOR2]] also possesses 7 [[transmembrane domain]]s. However ADIPOR1 and ADIPOR2 are orientated oppositely to GPCRs in the membrane (i.e., cytoplasmic [[N-terminus]], extracellular [[C-terminus]]) and do not associate with [[G protein]]s.<ref name="pmid12802337">{{cite journal | author = Yamauchi T, Kamon J, Ito Y, Tsuchida A, Yokomizo T, Kita S, Sugiyama T, Miyagishi M, Hara K, Tsunoda M, Murakami K, Ohteki T, Uchida S, Takekawa S, Waki H, Tsuno NH, Shibata Y, Terauchi Y, Froguel P, Tobe K, Koyasu S, Taira K, Kitamura T, Shimizu T, Nagai R, Kadowaki T | title = Cloning of adiponectin receptors that mediate antidiabetic metabolic effects | journal = Nature | volume = 423 | issue = 6941 | pages = 762–9 | year = 2003 | month = June | pmid = 12802337 | doi = 10.1038/nature01705 }}</ref> |
|||
Early structural models for GPCRs were based on their weak analogy to [[bacteriorhodopsin]], for which a structure had been determined by both electron diffraction ({{PDB|2BRD}}, {{PDB2|1AT9}})<ref name="pmid8676377">{{cite journal | author = Grigorieff N, Ceska TA, Downing KH, Baldwin JM, Henderson R | title = Electron-crystallographic refinement of the structure of bacteriorhodopsin | journal = J. Mol. Biol. | volume = 259 | issue = 3 | pages = 393–421 | year = 1996 | pmid = 8676377 | doi = 10.1006/jmbi.1996.0328 }}</ref><ref name="pmid9296502">{{cite journal | author = Kimura Y, Vassylyev DG, Miyazawa A, Kidera A, Matsushima M, Mitsuoka K, Murata K, Hirai T, Fujiyoshi Y | title = Surface of bacteriorhodopsin revealed by high-resolution electron crystallography | journal = Nature | volume = 389 | issue = 6647 | pages = 206–11 | year = 1997 | pmid = 9296502 | doi = 10.1038/38323 }}</ref> and [[X-ray crystallography|X ray-based crystallography]] ({{PDB2|1AP9}}).<ref name="pmid9287223">{{cite journal | author = Pebay-Peyroula E, Rummel G, Rosenbusch JP, Landau EM | title = X-ray structure of bacteriorhodopsin at 2.5 angstroms from microcrystals grown in lipidic cubic phases | journal = Science | volume = 277 | issue = 5332 | pages = 1676–81 | year = 1997 | pmid = 9287223 | doi = 10.1126/science.277.5332.1676 }}</ref> In 2000, the first crystal structure of a mammalian GPCR, that of bovine [[rhodopsin]] ({{PDB2|1F88}}), was solved.<ref name="pmid10926528">{{cite journal | author = Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Trong IL, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M | title = Crystal structure of rhodopsin: A G protein-coupled receptor | journal = Science | volume = 289 | issue = 5480 | pages = 739–45 | year = 2000 | pmid = 10926528 | doi = 10.1126/science.289.5480.739 }}</ref> While the main feature, the seven transmembrane helices, is conserved, the relative orientation of the helices differ significantly from that of bacteriorhodopsin. In 2007, the first structure of a human GPCR was solved ({{PDB2|2R4R}}, {{PDB2|2R4S}}).<ref name="Rasmussen_2007">{{cite journal | author = Rasmussen SG, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VR, Sanishvili R, Fischetti RF, Schertler GF, Weis WI, Kobilka BK | title = Crystal structure of the human β<sub>2</sub>-adrenergic G-protein-coupled receptor | journal = Nature | volume = 450 | issue = 7168 | pages = 383–7 | year = 2007 | pmid = 17952055 | doi = 10.1038/nature06325 }}</ref> This was followed immediately by a higher resolution structure of the same receptor ({{PDB2|2RH1}}).<ref name="Cherezov_2007">{{cite journal | author = Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC | title = High-resolution crystal structure of an engineered human β<sub>2</sub>-adrenergic G protein-coupled receptor | journal = Science | volume = 318 | issue = 5854 | pages = 1258–65 | year = 2007 | pmid = 17962520 | doi = 10.1126/science.1150577| pmc = 2583103 }}</ref><ref name="Rosenbaum_2007">{{cite journal | author = Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Yao XJ, Weis WI, Stevens RC, Kobilka BK | title = GPCR engineering yields high-resolution structural insights into β<sub>2</sub>-adrenergic receptor function | journal = Science | volume = 318 | issue = 5854 | pages = 1266–73 | year = 2007 | pmid = 17962519 | doi = 10.1126/science.1150609 }}</ref> This human [[beta-2 adrenergic receptor|β<sub>2</sub>-adrenergic receptor]] GPCR structure, proved to be highly similar to the bovine rhodopsin in terms of the relative orientation of the seven-transmembrane helices. However the conformation of the second extracellular loop is entirely different between the two structures. Since this loop constitutes the "lid" that covers the top of the ligand binding site, this conformational difference highlights the difficulties in constructing [[homology modeling|homology model]]s of other GPCRs based only on the rhodopsin structure. |
Early structural models for GPCRs were based on their weak analogy to [[bacteriorhodopsin]], for which a structure had been determined by both electron diffraction ({{PDB|2BRD}}, {{PDB2|1AT9}})<ref name="pmid8676377">{{cite journal | author = Grigorieff N, Ceska TA, Downing KH, Baldwin JM, Henderson R | title = Electron-crystallographic refinement of the structure of bacteriorhodopsin | journal = J. Mol. Biol. | volume = 259 | issue = 3 | pages = 393–421 | year = 1996 | pmid = 8676377 | doi = 10.1006/jmbi.1996.0328 }}</ref><ref name="pmid9296502">{{cite journal | author = Kimura Y, Vassylyev DG, Miyazawa A, Kidera A, Matsushima M, Mitsuoka K, Murata K, Hirai T, Fujiyoshi Y | title = Surface of bacteriorhodopsin revealed by high-resolution electron crystallography | journal = Nature | volume = 389 | issue = 6647 | pages = 206–11 | year = 1997 | pmid = 9296502 | doi = 10.1038/38323 }}</ref> and [[X-ray crystallography|X ray-based crystallography]] ({{PDB2|1AP9}}).<ref name="pmid9287223">{{cite journal | author = Pebay-Peyroula E, Rummel G, Rosenbusch JP, Landau EM | title = X-ray structure of bacteriorhodopsin at 2.5 angstroms from microcrystals grown in lipidic cubic phases | journal = Science | volume = 277 | issue = 5332 | pages = 1676–81 | year = 1997 | pmid = 9287223 | doi = 10.1126/science.277.5332.1676 }}</ref> In 2000, the first crystal structure of a mammalian GPCR, that of bovine [[rhodopsin]] ({{PDB2|1F88}}), was solved.<ref name="pmid10926528">{{cite journal | author = Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Trong IL, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M | title = Crystal structure of rhodopsin: A G protein-coupled receptor | journal = Science | volume = 289 | issue = 5480 | pages = 739–45 | year = 2000 | pmid = 10926528 | doi = 10.1126/science.289.5480.739 }}</ref> While the main feature, the seven transmembrane helices, is conserved, the relative orientation of the helices differ significantly from that of bacteriorhodopsin. In 2007, the first structure of a human GPCR was solved ({{PDB2|2R4R}}, {{PDB2|2R4S}}).<ref name="Rasmussen_2007">{{cite journal | author = Rasmussen SG, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VR, Sanishvili R, Fischetti RF, Schertler GF, Weis WI, Kobilka BK | title = Crystal structure of the human β<sub>2</sub>-adrenergic G-protein-coupled receptor | journal = Nature | volume = 450 | issue = 7168 | pages = 383–7 | year = 2007 | pmid = 17952055 | doi = 10.1038/nature06325 }}</ref> This was followed immediately by a higher resolution structure of the same receptor ({{PDB2|2RH1}}).<ref name="Cherezov_2007">{{cite journal | author = Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC | title = High-resolution crystal structure of an engineered human β<sub>2</sub>-adrenergic G protein-coupled receptor | journal = Science | volume = 318 | issue = 5854 | pages = 1258–65 | year = 2007 | pmid = 17962520 | doi = 10.1126/science.1150577| pmc = 2583103 }}</ref><ref name="Rosenbaum_2007">{{cite journal | author = Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Yao XJ, Weis WI, Stevens RC, Kobilka BK | title = GPCR engineering yields high-resolution structural insights into β<sub>2</sub>-adrenergic receptor function | journal = Science | volume = 318 | issue = 5854 | pages = 1266–73 | year = 2007 | pmid = 17962519 | doi = 10.1126/science.1150609 }}</ref> This human [[beta-2 adrenergic receptor|β<sub>2</sub>-adrenergic receptor]] GPCR structure, proved to be highly similar to the bovine rhodopsin in terms of the relative orientation of the seven-transmembrane helices. However the conformation of the second extracellular loop is entirely different between the two structures. Since this loop constitutes the "lid" that covers the top of the ligand binding site, this conformational difference highlights the difficulties in constructing [[homology modeling|homology model]]s of other GPCRs based only on the rhodopsin structure. |
||
Revision as of 03:09, 12 June 2012
| GPCR | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 | |||||||||
| Identifiers | |||||||||
| Symbol | 7tm_1 | ||||||||
| Pfam | PF00001 | ||||||||
| InterPro | IPR000276 | ||||||||
| PROSITE | PDOC00210 | ||||||||
| OPM superfamily | 6 | ||||||||
| OPM protein | 1gzm | ||||||||
| |||||||||

G protein coupled receptors (GPCRs), also known as seven-transmembrane domain receptors, 7TM receptors, heptahelical receptors, serpentine receptor, and G protein-linked receptors (GPLR), comprise a large protein family of transmembrane receptors that sense molecules outside the cell and activate inside signal transduction pathways and, ultimately, cellular responses. G protein-coupled receptors are found only in eukaryotes, including yeast, choanoflagellates,[2] and animals. The ligands that bind and activate these receptors include light-sensitive compounds, odors, pheromones, hormones, and neurotransmitters, and vary in size from small molecules to peptides to large proteins. G protein-coupled receptors are involved in many diseases, and are also the target of approximately 40% of all modern medicinal drugs.[3][4]
There are two principal signal transduction pathways involving the G protein-coupled receptors: the cAMP signal pathway and the phosphatidylinositol signal pathway.[5] When a ligand binds to the GPCR it causes a conformational change in the GPCR, which allows it to act as a guanine nucleotide exchange factor (GEF). The GPCR can then activate an associated G-protein by exchanging its bound GDP for a GTP. The G-protein's α subunit, together with the bound GTP, can then dissociate from the β and γ subunits to further affect intracellular signaling proteins or target functional proteins directly depending on the α subunit type (Gαs, Gαi/o, Gαq/11, Gα12/13).[6]: 1160
Classification

The exact size of the GPCR superfamily is unknown but nearly 800 different human genes (or ≈4% of the entire protein-coding genome) have been predicted from genome sequence analysis. Although numerous classification schemes have been proposed, the superfamily is classically divided into three main classes (A, B, and C) with no detectable shared sequence homology between classes. The largest class by far is class A, which accounts for nearly 85% of the GPCR genes. Of class A GPCRs, over half of these are predicted to encode olfactory receptors while the remaining receptors are liganded by known endogenous compounds or are classified as orphan receptors. Despite the lack of sequence homology between classes, all GPCRs share a common structure and mechanism of signal transduction.
In all, GPCRs can be grouped into 6 classes based on sequence homology and functional similarity:[8][9][10][11]
- Class A (or 1) (Rhodopsin-like)
- Class B (or 2) (Secretin receptor family)
- Class C (or 3) (Metabotropic glutamate/pheromone)
- Class D (or 4) (Fungal mating pheromone receptors)
- Class E (or 5) (Cyclic AMP receptors)
- Class F (or 6) (Frizzled/Smoothened)
The very large rhodopsin A group has been further subdivided into 19 subgroups (A1-A19).[12] More recently, an alternative classification system called GRAFS (Glutamate, Rhodopsin, Adhesion, Frizzled/Taste2, Secretin) has been proposed.[13]
The human genome encodes thousands of G protein-coupled receptors,[14] about 350 of which detect hormones, growth factors, and other endogenous ligands. Approximately 150 of the GPCRs found in the human genome have unknown functions.
Some web-servers[15] and bioinformatics prediction methods[16][17] have been used for predicting the classification of GPCRs according to their amino acid sequence alone, by means of the pseudo amino acid composition approach.
Physiological roles
GPCRs are involved in a wide variety of physiological processes. Some examples of their physiological roles include:
- The visual sense: the opsins use a photoisomerization reaction to translate electromagnetic radiation into cellular signals. Rhodopsin, for example, uses the conversion of 11-cis-retinal to all-trans-retinal for this purpose
- The sense of smell: receptors of the olfactory epithelium bind odorants (olfactory receptors) and pheromones (vomeronasal receptors)
- Behavioral and mood regulation: receptors in the mammalian brain bind several different neurotransmitters, including serotonin, dopamine, GABA, and glutamate
- Regulation of immune system activity and inflammation: chemokine receptors bind ligands that mediate intercellular communication between cells of the immune system; receptors such as histamine receptors bind inflammatory mediators and engage target cell types in the inflammatory response
- Autonomic nervous system transmission: both the sympathetic and parasympathetic nervous systems are regulated by GPCR pathways, responsible for control of many automatic functions of the body such as blood pressure, heart rate, and digestive processes
- Cell density sensing: A novel GPCR role in regulating cell density sensing.
- Homeostasis modulation (e.g., water balance).[18]
Receptor structure
GPCRs are integral membrane proteins that possess seven membrane-spanning domains or transmembrane helices. The extracellular parts of the receptor can be glycosylated. These extracellular loops also contain two highly-conserved cysteine residues that form disulfide bonds to stabilize the receptor structure. Some seven-transmembrane helix proteins (channelrhodopsin) that resemble GPCRs may contain ion channels, within their protein.
Similar to (GPCRs, the adiponectin receptors 1 and 2 (ADIPOR1 and ADIPOR2 also possesses 7 transmembrane domains. However ADIPOR1 and ADIPOR2 are orientated oppositely to GPCRs in the membrane (i.e., cytoplasmic N-terminus, extracellular C-terminus) and do not associate with G proteins.[19]
Early structural models for GPCRs were based on their weak analogy to bacteriorhodopsin, for which a structure had been determined by both electron diffraction (PDB: 2BRD, 1AT9)[20][21] and X ray-based crystallography (1AP9).[22] In 2000, the first crystal structure of a mammalian GPCR, that of bovine rhodopsin (1F88), was solved.[23] While the main feature, the seven transmembrane helices, is conserved, the relative orientation of the helices differ significantly from that of bacteriorhodopsin. In 2007, the first structure of a human GPCR was solved (2R4R, 2R4S).[24] This was followed immediately by a higher resolution structure of the same receptor (2RH1).[25][26] This human β2-adrenergic receptor GPCR structure, proved to be highly similar to the bovine rhodopsin in terms of the relative orientation of the seven-transmembrane helices. However the conformation of the second extracellular loop is entirely different between the two structures. Since this loop constitutes the "lid" that covers the top of the ligand binding site, this conformational difference highlights the difficulties in constructing homology models of other GPCRs based only on the rhodopsin structure.
The structures of activated and/or agonist-bound GPCRs have also been determined.[27][28][29][30] These structures indicate how ligand binding at the extracellular side of a receptor leads to conformational changes in the cytoplasmic side of the receptor. The biggest change is an outward movement of the cytoplasmic part of the 5th and 6th transmembrane helix (TM5 and TM6). The structure of activated beta-2 adrenergic receptor in complex with Gs confirmed that the Gα binds to a cavity created by this movement.[31]
Structure-function relationships
Structurally, GPCRs are characterized by an extracellular N-terminus, followed by seven transmembrane (7-TM) α-helices (TM-1 to TM-7) connected by three intracellular (IL-1 to IL-3) and three extracellular loops (EL-1 to EL-3), and finally an intracellular C-terminus. The GPCR arranges itself into a tertiary structure resembling a barrel, with the seven transmembrane helices forming a cavity within the plasma membrane that serves a ligand-binding domain that is often covered by EL-2. Ligands may also bind elsewhere, however, as is the case for bulkier ligands (e.g., proteins or large peptides), which instead interact with the extracellular loops, or, as illustrated by the class C metabotropic glutamate receptors (mGluRs), the N-terminal tail. The class C GPCRs are distinguished by their large N-terminal tail, which also contains a ligand-binding domain. Upon glutamate-binding to an mGluR, the N-terminal tail undergoes a conformational change that leads to its interaction with the residues of the extracellular loops and TM domains. The eventual effect of all three types of agonist-induced activation is a change in the relative orientations of the TM helices (likened to a twisting motion) leading to a wider intracellular surface and "revelation" of residues of the intracellular helices and TM domains crucial to signal transduction function (i.e., G-protein coupling). Inverse agonists and antagonists may also bind to a number of different sites, but the eventual effect must be prevention of this TM helix reorientation.
The structure of the N- and C-terminal tails of GPCRs may also serve important functions beyond ligand-binding. In particular, the C-terminus often contains serine (Ser) or threonine (Thr) residues that, when phosphorylated, increase the affinity of the intracellular surface for the binding of scaffolding proteins called β-arrestins (β-arr).[32] Once bound, β-arrestins both sterically prevent G-protein coupling and may recruit other proteins leading to the creation of signaling complexes involved in extracellular-signal regulated kinase (ERK) pathway activation or receptor endocytosis (internalization). As the phosphorylation of these Ser and Thr residues often occurs as a result of GPCR activation, the β-arr-mediated G-protein-decoupling and internalization of GPCRs are important mechanisms of desensitization.[33]
A final common structural theme among GPCRs is palmitoylation of one or more sites of the C-terminal tail or the intracellular loops. Palmitoylation is the covalent modification of cysteine (Cys) residues via addition of hydrophobic acyl groups, and has the effect of targeting the receptor to cholesterol- and sphingolipid-rich microdomains of the plasma membrane called lipid rafts. As many of the downstream transducer and effector molecules of GPCRs (including those involved in negative feedback pathways) are also targeted to lipid rafts, this has the effect of facilitating rapid receptor signaling.
GPCRs respond to extracellular signals mediated by a huge diversity of agonists, ranging from proteins to biogenic amines to protons, but all transduce this signal via a mechanism of G-protein coupling. This is made possible by virtue of a guanine-nucleotide exchange factor (GEF) domain primarily formed by a combination of IL-2 and IL-3 along with adjacent residues of the associated TM helices.
Mechanism
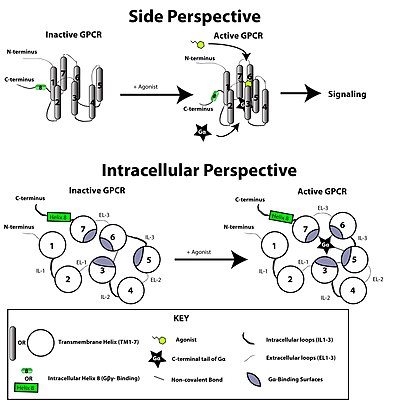
The G protein-coupled receptor is activated by an external signal in the form of a ligand or other signal mediator. This creates a conformational change in the receptor, causing activation of a G protein. Further effect depends on the type of G protein.
Ligand binding
GPCRs include receptors for sensory signal mediators (e.g., light and olfactory stimulatory molecules); adenosine, bombesin, bradykinin, endothelin, γ-aminobutyric acid (GABA), hepatocyte growth factor (HGF), melanocortins, neuropeptide Y, opioid peptides, opsins, somatostatin, GH, tachykinins, members of the vasoactive intestinal peptide family, and vasopressin; biogenic amines (e.g., dopamine, epinephrine, norepinephrine, histamine, glutamate (metabotropic effect), glucagon, acetylcholine (muscarinic effect), and serotonin); chemokines; lipid mediators of inflammation (e.g., prostaglandins, prostanoids, platelet-activating factor, and leukotrienes); and peptide hormones (e.g., calcitonin, C5a anaphylatoxin, follicle-stimulating hormone (FSH), gonadotropin-releasing hormone (GnRH), neurokinin, thyrotropin-releasing hormone (TRH), cannabinoids, and oxytocin). GPCRs that act as receptors for stimuli that have not yet been identified are known as orphan receptors.
Whereas, in other types of receptors that have been studied, wherein ligands bind externally to the membrane, the ligands of GPCRs typically bind within the transmembrane domain. However, protease-activated receptors are activated by cleavage of part of their extracellular domain.[35]
Conformational change

The transduction of the signal through the membrane by the receptor is not completely understood. It is known that the inactive G protein is bound to the receptor in its inactive state. Once the ligand is recognized, the receptor shifts conformation and, thus, mechanically activates the G protein, which detaches from the receptor. The receptor can now either activate another G protein or switch back to its inactive state. This is an overly simplistic explanation, but suffices to convey the overall set of events.
It is believed that a receptor molecule exists in a conformational equilibrium between active and inactive biophysical states.[36] The binding of ligands to the receptor may shift the equilibrium toward the active receptor states.[37] Three types of ligands exist: agonists are ligands that shift the equilibrium in favour of active states; inverse agonists are ligands that shift the equilibrium in favour of inactive states; and neutral antagonists are ligands that do not affect the equilibrium. It is not yet known how exactly the active and inactive states differ from each other.
G-protein activation/deactivation cycle

When the receptor is inactive, the GEF domain may be bound to an also inactive α-subunit of a heterotrimeric G-protein. These "G-proteins" are a trimer of α, β, and γ subunits (known as Gα, Gβ, and Gγ, respectively) that is rendered inactive when reversibly bound to Guanosine diphosphate (GDP) (or alternatively, no guanine nucleotide) but active when bound to Guanosine triphosphate (GTP). Upon receptor activation, the GEF domain, in turn, allosterically activates the G-protein by facilitating the exchange of a molecule of GDP for GTP at the G-protein's α-subunit. The cell maintains a 10:1 ratio of cytosolic GTP:GDP so exchange for GTP is ensured. At this point, the subunits of the G-protein dissociate from the receptor, as well as each other, to yield a Gα-GTP monomer and a tightly interacting Gβγ dimer, which are now free to modulate the activity of other intracellular proteins. The extent to which they may diffuse, however, is limited due to the palmitoylation of Gα and the presence of a molecule of Glycosylphosphatidylinositol (GPI) that has been covalently added to the C-termini of Gγ. The phosphatidylinositol moiety of the GPI-linkage contains two hydrophobic acyl groups that anchor any GPI-linked proteins (e.g. Gβγ) to the plasma membrane, and also, to some extent, to the local lipid raft. (Compare this to the effect of palmitoylation on GPCR localization discussed above)
Because Gα also has slow GTP→GDP hydrolysis capability, the inactive form of the α-subunit (Gα-GDP) is eventually regenerated, thus allowing reassociation with a Gβγ dimer to form the "resting" G-protein, which can again bind to a GPCR and await activation. The rate of GTP hydrolysis is often accelerated due to the actions of another family of allosteric modulating proteins called Regulators of G-protein Signaling, or RGS proteins, which are a type of GTPase-Activating Protein, or GAP. In fact, many of the primary effector proteins (e.g. adenylate cyclases) that become activated/inactivated upon interaction with Gα-GTP also have GAP activity. Thus, even at this early stage in the process, GPCR-initiated signaling has the capacity for self-termination.
GPCR signaling

If a receptor in an active state encounters a G protein, it may activate it. Some evidence suggests that receptors and G proteins are actually pre-coupled. For example, binding of G proteins to receptors affects the receptor's affinity for ligands. Activated G proteins are bound to GTP.
Further signal transduction depends on the type of G protein. The enzyme adenylate cyclase is an example of a cellular protein that can be regulated by a G protein, in this case the G protein Gs. Adenylate cyclase activity is activated when it binds to a subunit of the activated G protein. Activation of adenylate cyclase ends when the G protein returns to the GDP-bound state.
Adenylate cyclases (of which 9 membrane-bound and one cytosolic forms are known in humans) may also be activated or inhibited in other ways (e.g., Ca2+/Calmodulin binding), which can modify the activity of these enzymes in an additive or synergistic fashion along with the G proteins.
The signaling pathways activated through a GPCR are limited by the primary sequence and tertiary structure of the GPCR itself but ultimately determined by the particular conformation stabilized by a particular ligand, as well as the availability of transducer molecules. Currently, GPCRs are considered to utilize two primary types of transducers: G-proteins and β-arrestins. Because β-arr’s only have high affinity to the phosphorylated form of most GPCRs (see above or below), the majority of signaling is ultimately dependent upon G-protein activation. However, the possibility for interaction does allow for G-protein independent signaling to occur.
G-protein-dependent signaling
There are three main G-protein-mediated signaling pathways, mediated by four sub-classes of G-proteins distinguished from each other by sequence homology (Gαs, Gαi/o, Gαq/11, and Gα12/13). Each sub-class of G-protein consists of multiple proteins, each the product of multiple genes and/or splice variations that may imbue them with differences ranging from subtle to distinct with regard to signaling properties, but in general they appear to be reasonably grouped into four classes. Because the signal transducing properties of the various possible βγ combinations do not appear to radically differ from one another, these classes are defined according to the isoform of their α-subunit.[6]: 1163
While most GPCRs are capable of activating more than one Gα-subtype, they also show a preference for one subtype over another. When the subtype activated depends on the ligand that is bound to the GPCR, this is called functional selectivity (also known as agonist-directed trafficking, or conformation specific agonism). However, the binding of any single particular agonist may also initiate activation of multiple different G-proteins, as it may be capable of stabilizing more than one conformation of the GPCR’s GEF domain, even over the course of a single interaction. Additionally, a conformation that preferably activates one isoform of Gα may activate another if the preferred is less available. Furthermore, feedback pathways may result in receptor modifications (e.g. phosphorylation) that alter the G-protein preference. Regardless of these various nuances, the GPCR’s preferred coupling partner is usually defined according to the G-protein most obviously activated by the endogenous ligand under most physiological and/or experimental conditions.
Gα signaling
- The effector of both the Gαs and Gαi/o pathways is the Cyclic-adenosine monophosphate (cAMP) generating enzyme Adenylate Cyclase, or AC. While there are ten different AC gene products in mammals, each with subtle differences in tissue distribution and/or function, all catalyze the conversion of cytosolic Adenosine Triphosphate (ATP) to cAMP, and all are directly stimulated by G-proteins of the Gαs class. Conversely, interaction with Gα subunits of the Gαi/o type inhibits AC from generating cAMP. Thus, a GPCR coupled to Gαs will counteract the actions of a GPCR coupled to Gαi/o, and vice versa. The level of cytosolic cAMP may then determine the activity of various ion channels as well as members of the ser/thr specific Protein Kinase A (PKA) family. Thus cAMP is considered a second messenger and PKA a secondary effector.
- The effector of the Gαq/11 pathway is Phospholipase C-β (PLCβ), which catalyzes the cleavage of membrane-bound phosphatidylinositol 4,5-biphosphate (PIP2) into the second messengers inositol (1,4,5) trisphosphate (IP3) and diacylglycerol (DAG). IP3 acts on IP3 receptors found in the membrane of the endoplasmic reticulum (ER) to elicit Ca2+ release from the ER, while DAG diffuses along the plasma membrane where it may activate any membrane localized forms of a second ser/thr kinase called Protein Kinase C (PKC). Since many isoforms of PKC are also activated by increases in intracellular Ca2+, both these pathways can also converge on each other to signal through the same secondary effector. Elevated intracellular Ca2+ also binds and allosterically activates proteins called Calmodulins, which in turn go on to bind and allosterically activate enzymes such as Ca2+/Calmodulin-dependant Kinases (CAMKs).
- The effectors of the Gα12/13 pathway are three RhoGEFs (p115-RhoGEF, PDZ-RhoGEF, and LARG), which, when bound to Gα12/13 allosterically activate the cytosolic small GTPase, Rho. Once bound to GTP, Rho can then go on to activate various proteins responsible for cytoskeleton regulation such as Rho-kinase (ROCK). Most GPCRs that couple to Gα12/13 also couple to other sub-classes, often Gαq/11.
Gβγ signaling
The above descriptions ignore the effects of Gβγ–signalling, which can also be important, in particular in the case of activated Gαi/o-coupled GPCRs. The primary effectors of Gβγ are various ion channels, such as G-protein-regulated Inwardly Rectifying K+ channels (GIRKs), P/Q- and N-type voltage-gated Ca2+ Channels, as well as some isoforms of AC and PLC, along with some Phosphoinositide-3-Kinase (PI3K) isoforms.
G-Protein-independent signaling
Although they are classically thought of working only together, GPCRs may signal through G-protein-independent mechanisms, and heterotrimeric G-proteins may play functional roles independent of GPCRs. GPCRs may signal independently through many proteins already mentioned for their roles in G-protein-dependent signaling such as β-arrs, GRKs, and Srcs. Additionally, further scaffolding proteins involved in subcellular localization of GPCRs (e.g., PDZ-domain-containing proteins) may also act as signal transducers. Most often the effector is a member of the MAPK family.
Examples
In the late 1990s, evidence began accumulating to suggest that some GPCRs are able to signal without G proteins. The ERK2 mitogen-activated protein kinase, a key signal transduction mediator downstream of receptor activation in many pathways, has been shown to be activated in response to cAMP-mediated receptor activation in the slime mold D. discoideum despite the absence of the associated G protein α- and β-subunits.[38]
In mammalian cells, the much-studied β2-adrenoceptor has been demonstrated to activate the ERK2 pathway after arrestin-mediated uncoupling of G-protein-mediated signaling. Therefore it seems likely that some mechanisms previously believed to be purely related to receptor desensitisation are actually examples of receptors switching their signaling pathway rather than simply being switched off.
In kidney cells, the bradykinin receptor B2 has been shown to interact directly with a protein tyrosine phosphatase. The presence of a tyrosine-phosphorylated ITIM (immunoreceptor tyrosine-based inhibitory motif) sequence in the B2 receptor is necessary to mediate this interaction and subsequently the antiproliferative effect of bradykinin.[39]
GPCR-independent signaling by heterotrimeric G-proteins
Although it is a relatively immature area of research, it appears that heterotrimeric G-proteins may also take part in non-GPCR signaling. There is evidence for roles as signal transducers in nearly all other types of receptor-mediated signaling, including integrins, receptor tyrosine kinases (RTKs), cytokine receptors (JAK/STATs), as well as modulation of various other "accessory" proteins such as GEFs, Guanine-nucleotide Dissociation Inhibitors (GDIs) and protein phosphatases. There may even be specific proteins of these classes whose primary function is as part of GPCR-independent pathways, termed Activators of G-protein Signalling (AGS). Both the ubiquity of these interactions and the importance of Gα vs. Gβγ subunits to these processes are still unclear.
Details of cAMP and PIP2 pathways



There are two principal signal transduction pathways involving the G protein-linked receptors: cAMP signal pathway and Phosphatidylinositol signal pathway.[5]
cAMP signal pathway
The cAMP signal transduction contains 5 main characters: stimulative hormone receptor (Rs) or inhibitory hormone receptor (Ri);Stimulative regulative G-protein (Gs) or inhibitory regulative G-protein (Gi);Adenylyl cyclase; Protein Kinase A (PKA); and cAMP phosphodiesterase.
Stimulative hormone receptor (Rs) is a receptor that can bind with stimulative signal molecules, while inhibitory hormone (Ri) is a receptor that can bind with inhibitory signal molecules.
Stimulative regulative G-protein is a G protein-linked to stimulative hormone receptor (Rs) and its α subunit upon activation could stimulate the activity of an enzyme or other intracellular metabolism. On the contrary, inhibitory regulative G-protein is linked to an inhibitory hormone receptor and its α subunit upon activation could inhibit the activity of an enzyme or other intracellular metabolism.
The Adenylyl cyclase is a 12-transmembrane glucoprotein that catalyzes ATP to form cAMP with the help of cofactor Mg2+ or Mn2+. The cAMP produced is a second messenger in cellular metabolism and is an allosteric activator to Protein kinase A.
Protein kinase A is an important enzyme in cell metabolism due to its ability to regulate cell metabolism by phosphorylating specific committed enzymes in the metabolic pathway. It can also regulate specific gene expression, cellular secretion, and membrane permeability. The protein enzyme contains two catalytic subunits and two regulatory subunits. When there is no cAMP,the complex is inactive. When cAMP binds to the regulatory subunits, their conformation is altered, causing the dissociation of the regulatory subunits, which activates protein kinase A and allows further biological effects.
cAMP phosphodiesterase is an enzyme that can degrade cAMP to 5'-AMP, which will terminate the signal.
Phosphatidylinositol signal pathway
In the phosphatidylinositol signal pathway, the extracellular signal molecule binds with the G-protein receptor (Gq) on the cell surface and activates phospholipase C, which is located on the plasma membrane. The lipase hydrolyzes phosphatidylinositol 4,5-bisphosphate (PIP2) into two second messengers: Inositol 1,4,5-trisphosphate (IP3) and Diacylglycerol (DAG). IP3 binds with the receptor in the membrane of the smooth endoplasmic reticulum and mitochondria, help open the Ca2+ channel. DAG will help activate Protein Kinase C (PKC), which phosphorylates many other proteins, changing their catalytic activities, leading to cellular responses. The effects of Ca2+ is also remarkable: it cooperates with DAG in activating PKC and can activate CaM kinase pathway, in which calcium modulated protein calmodulin (CaM) binds Ca2+, undergoes a change in conformation, and activates CaM kinase II, which has unique ability to increase its binding affinity to CaM by autophosphorylation, making CaM unavailable for the activation of other enzymes. The kinase then phosphorylates target enzymes, regulating their activities. The two signal pathways are connected together by Ca2+-CaM, which is also a regulatory subunit of adenylyl cyclase and phosphodiesterase in cAMP signal pathway.
Receptor regulation
GPCRs become desensitized when exposed to their ligand for a prolonged period of time. There are two recognized forms of desensitization: 1) homologous desensitization, in which the activated GPCR is downregulated; and 2) heterologous desensitization, wherein the activated GPCR causes downregulation of a different GPCR. The key reaction of this downregulation is the phosphorylation of the intracellular (or cytoplasmic) receptor domain by protein kinases.
Phosphorylation by cAMP-dependent protein kinases
Cyclic AMP-dependent protein kinases (protein kinase A) are activated by the signal chain coming from the G protein (that was activated by the receptor) via adenylate cyclase and cyclic AMP (cAMP). In a feedback mechanism, these activated kinases phosphorylate the receptor. The longer the receptor remains active, the more kinases are activated, the more receptors are phosphorylated. In β2-adrenoceptors, this phosphorylation results in the switching of the coupling from the Gs class of G-protein to the Gi class.[40] cAMP-dependent PKA mediated phosphorylation can cause heterologous desensitisation in receptors other than those activated.[41]
Phosphorylation by GRKs
The G protein-coupled receptor kinases (GRKs) are protein kinases that phosphorylate only active GPCRs.
Phosphorylation of the receptor can have two consequences:
- Translocation: The receptor is, along with the part of the membrane it is embedded in, brought to the inside of the cell, where it is dephosphorylated within the acidic vesicular environment[42] and then brought back. This mechanism is used to regulate long-term exposure, for example, to a hormone, by allowing resensitisation to follow desensitisation. Alternatively, the receptor may undergo lysozomal degradation, or remain internalised, where it is thought to participate in the initiation of signalling events, the nature of which depend on the internalised vesicle's subcellular localisation.[41]
- Arrestin linking: The phosphorylated receptor can be linked to arrestin molecules that prevent it from binding (and activating) G proteins, effectively switching it off for a short period of time. This mechanism is used, for example, with rhodopsin in retina cells to compensate for exposure to bright light. In many cases, arrestin binding to the receptor is a prerequisite for translocation. For example, beta-arrestin bound to β2-adrenoreceptors acts as an adaptor for binding with clathrin, and with the beta-subunit of AP2 (clathrin adaptor molecules); thus the arrestin here acts as a scaffold assembling the components needed for clathrin-mediated endocytosis of β2-adrenoreceptors.[43][44]
Mechanisms of GPCR signal termination
As mentioned above, G-proteins may terminate their own activation due to their intrinsic GTP→GDP hydrolysis capability. However, this reaction proceeds at a slow rate (≈.02 times/sec) and thus it would take around 50 seconds for any single G-protein to deactivate if other factors did not come into play. Indeed, there are around 30 isoforms of RGS proteins that, when bound to Gα through their GAP domain, accelerate the hydrolysis rate to ≈30 times/sec. This 1500-fold increase in rate allows for the cell to respond to external signals with high speed, as well as spatial resolution due to limited amount of second messenger that can be generated and limited distance a G-protein can diffuse in .03 seconds. For the most part, the RGS proteins are promiscuous in their ability to activate G-proteins, while which RGS is involved in a given signaling pathway seems to be more determined by the tissue and GPCR involved than anything else. Additionally, RGS proteins have the additional function of increasing the rate of GTP-GDP exchange at GPCRs, (i.e. as a sort of co-GEF) further contributing to the time resolution of GPCR signaling.
In addition, the GPCR may be desensitized itself. This can occur as:
- a direct result of ligand occupation, wherein the change in conformation allows recruitment of GPCR-Regulating Kinases (GRKs), which go on to phosphorylate various serine/threonine residues of IL-3 and the C-terminal tail. Upon GRK phosphorylation, the GPCR's affinity for β-arrestin (β-arrestin-1/2 in most tissues) is increased, at which point β-arrestin may bind and act to both sterically hinder G-protein coupling as well as initiate the process of receptor internalization through clathrin-mediated endocytosis. Because only the liganded receptor is desensitized by this mechanism, it is called homologous desensitization
- the affinity for β-arr may increased in a ligand occupation and GRK-independent manner through phosphorylation of different ser/thr sites (but also of IL-3 and the C-terminal tail) by PKC and PKA. These phosphorylations are often sufficient to impair G-protein coupling on their own as well.
- PKC/PKA may, instead, phosphorylate GRKs, which can also lead to GPCR phosphorylation and β-arrestin binding in an occupation-independent manner. These latter two mechanisms allow for desensitization of one GPCR due to the activities of others, or heterologous desensitization. GRKs may also have GAP domains and so may contribute to inactivation through non-kinase mechanisms as well. A combination of these mechanisms may also occur.
Once β-arrestin is bound to a GPCR, it undergoes a conformational change allowing it to serve as a scaffolding protein for an adaptor complex termed AP-2, which in turn recruits another protein called clathrin. If enough receptors in the local area recruit clathrin in this manner, they aggregate and the membrane buds inwardly as a result of interactions between the molecules of clathrin, in a process called opsonization. Once the pit has been pinched off, the plasma membrane due to the actions of two other proteins called amphiphysin and dynamin, it is now an endocytic vesicle. At this point the adapter molecules and clathrin have dissociated and the receptor will be either trafficked back to the plasma membrane or targeted to lysosomes for degradation.
At any point in this process, the β-arrestins may also be recruiting other proteins such as the non-receptor tyrosine kinase (nRTK), c-SRC, which may initiate activation of ERK1/2, or other mitogen-activated protein kinase (MAPK) signaling through, for example, phosphorylation of the small GTP-ase, Ras, or recruit the proteins of the ERK cascade directly (i.e., Raf-1, MEK, ERK-1/2) at which point signaling is initiated due to their close proximity to one another. Another target of c-SRC are the dynamin molecules involved in endocytosis. Dynamins polymerize around the neck of an incoming vesicle, and their phosphorylation by c-SRC provides the energy necessary for the conformational change allowing the final "pinching off" from the membrane.
GPCR cellular regulation
Receptor desensitization is mediated through a combination phosphorylation, β-arr binding, and endocytosis as described above. Downregulation occurs when endocytosed receptor is embedded in an endosome that is trafficked to merge with an organelle called a lysosome. Because lysosomal membranes are rich in proton pumps, their interiors have low pH (≈4.8 vs. the pH≈7.2 cytosol), which acts to denature the GPCRs. Additionally, lysosomes contain many degradative enzymes, including proteases, which can function only at such low pH, and so the peptide bonds joining the residues of the GPCR together may be cleaved. Whether or not a given receptor is trafficked to a lysosome, detained in endosomes, or trafficked back to the plasma membrane depends on a variety of factors, including receptor type and magnitude of the signal. GPCR regulation is additionally mediated by gene transcription factors. These factors can increase or decrease gene transcription and thus increase or decrease the generation of new receptors (up- or down-regulation) that will travel to the cell membrane.
Receptor oligomerization
G-protein-coupled receptor oligomerisation is a widespread phenomenon. One of the best-studied example is the metabotropic GABAB receptor. This so-called constitutive receptor is formed by heterodimerization of GABABR1 and GABABR2 subunits. Expression of the GABABR1 without the GABABR2 in heterologous systems leads to retention of the subunit in the endoplasmic reticulum. Expression of the GABABR2 subunit alone, meanwhile, leads to surface expression of the subunit, although with no functional activity (i.e., the receptor does not bind agonist and cannot initiate a response following exposure to agonist). Expression of the two subunits together leads to plasma membrane expression of functional receptor. It has been shown that GABABR2 binding to GABABR1 causes masking of a retention signal[45] of functional receptors.[46]
The Origin and diversification of the Superfamily
The signal transduction mediated by the superfamily of GPCRs were traced back to the origin of multicellularity. Recent reports have shown that the mammalian like GPCRs were found in the Fungi Kingdom and were classified according to the GRAFS classification system based on the GPCR fingerprints.[47] Identification of the superfamily members across the Eukaryotic domain and comparison of the family specific motifs have shown that the superfamily of GPCRs have a common origin.[48] Characteristic motifs support that three among the five GRAFS families, Rhodopsin, Adhesion and Frizzled evolved from the Dictyostelium discoideum cAMP receptors before the split of Opisthokonts. Later the Secretin family evolved from the Adhesion receptor family before the split of nematodes.
Dictyostelium discoideum
A novel GPCR containing a lipid kinase domain has recently been identified in Dictyostelium discoideum that regulates cell density sensing.[49]
See also
- Orphan receptor
- Pepducins, a class of drug candidates targeted at GPCRs
- G protein-coupled receptors database
- Metabotropic receptor
References
- ^ Huixian Wu, Daniel Wacker, Mauro Mileni, Vsevolod Katritch, GyeWon Han, Eyal Vardy, Wei Liu, Aaron A. Thompson, Xi-Ping Huang, F. Ivy Carroll, S. Wayne Mascarella, Richard B. Westkaemper, Philip D. Mosier, Bryan L. Roth, Vadim Cherezov & Raymond C. Stevens (2012). "Structure of the human k-opioid receptor in complex with JDTic". Nature. doi:10.1038/nature10939.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ King N, Hittinger CT, Carroll SB (2003). "Evolution of key cell signaling and adhesion protein families predates animal origins". Science. 301 (5631): 361–3. doi:10.1126/science.1083853. PMID 12869759.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Filmore, David (2004). "It's a GPCR world". Modern Drug Discovery. 2004 (November). American Chemical Society: 24–28.
- ^ Overington JP, Al-Lazikani B, Hopkins AL (2006). "How many drug targets are there?". Nat Rev Drug Discov. 5 (12): 993–6. doi:10.1038/nrd2199. PMID 17139284.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ a b Gilman A.G. (1987). "G Proteins: Transducers of Receptor-Generated Signals". Annual Review of Biochemistry. 56: 615–649. doi:10.1146/annurev.bi.56.070187.003151. PMID 3113327.
- ^ a b Wettschureck N, Offermanns S (2005). "Mammalian G proteins and their cell type specific functions". Physiol. Rev. 85 (4): 1159–204. doi:10.1152/physrev.00003.2005. PMID 16183910.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Bjarnadóttir TK, Gloriam DE, Hellstrand SH, Kristiansson H, Fredriksson R, Schiöth HB (2006). "Comprehensive repertoire and phylogenetic analysis of the G protein-coupled receptors in human and mouse". Genomics. 88 (3): 263–73. doi:10.1016/j.ygeno.2006.04.001. PMID 16753280.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Attwood TK, Findlay JB (1994). "Fingerprinting G-protein-coupled receptors". Protein Eng. 7 (2): 195–203. doi:10.1093/protein/7.2.195. PMID 8170923.
- ^ Kolakowski LF Jr (1994). "GCRDb: a G-protein-coupled receptor database". Receptors Channels. 2 (1): 1–7. PMID 8081729.
- ^ Foord SM, Bonner TI, Neubig RR, Rosser EM, Pin JP, Davenport AP, Spedding M, Harmar AJ (2005). "International Union of Pharmacology. XLVI. G protein-coupled receptor list". Pharmacol Rev. 57 (2): 279–88. doi:10.1124/pr.57.2.5. PMID 15914470.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ InterPro
- ^ Joost P, Methner A (2002). "Phylogenetic analysis of 277 human G-protein-coupled receptors as a tool for the prediction of orphan receptor ligands". Genome Biol. 3 (11): research0063.1–0063.16. doi:10.1186/gb-2002-3-11-research0063. PMC 133447. PMID 12429062.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Bjarnadottir TK, Gloriam DE, Hellstrand SH, Kristiansson H, Fredriksson R, Schioth HB (2006). "Comprehensive repertoire and phylogenetic analysis of the G protein-coupled receptors in human and mouse". Genomics. 88 (3): 263–73. doi:10.1016/j.ygeno.2006.04.001. PMID 16753280.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Vassilatis DK, Hohmann JG, Zeng H, Li F, Ranchalis JE; et al. (2003). "The G protein-coupled receptor repertoires of human and mouse". Proc Natl Acad Sci USA. 100 (8): 4903–4908. doi:10.1073/pnas.0230374100. PMC 153653. PMID 12679517.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Xiao X, Wang P, Chou KC (2009). "A cellular automaton image approach for predicting G-protein-coupled receptor functional classes". Journal of Computational Chemistry. 30 (9): 1414–1423. doi:10.1002/jcc.21163. PMID 19037861.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Qiu JD, Huang JH, Liang RP, Lu XQ (2009). "Prediction of G-protein-coupled receptor classes based on the concept of Chou's pseudo amino acid composition: an approach from discrete wavelet transform". Anal. Biochem. 390 (1): 68–73. doi:10.1016/j.ab.2009.04.009. PMID 19364489.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Gu Q, Ding YS, Zhang TL (2010). "Prediction of G-Protein-Coupled Receptor Classes in Low Homology Using Chou's pseudo amino acid composition with Approximate Entropy and Hydrophobicity Patterns". Protein Pept. Lett. 17 (5): 559–67. doi:10.2174/092986610791112693. PMID 19594431.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Hazell GG, Hindmarch CC, Pope GR, Roper JA, Lightman SL, Murphy D, O'Carroll AM, Lolait SJ (2011). "G protein-coupled receptors in the hypothalamic paraventricular and supraoptic nuclei - serpentine gateways to neuroendocrine homeostasis". Front Neuroendocrinol. 33 (1): 45–66. doi:10.1016/j.yfrne.2011.07.002. PMC 3336209. PMID 21802439.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Yamauchi T, Kamon J, Ito Y, Tsuchida A, Yokomizo T, Kita S, Sugiyama T, Miyagishi M, Hara K, Tsunoda M, Murakami K, Ohteki T, Uchida S, Takekawa S, Waki H, Tsuno NH, Shibata Y, Terauchi Y, Froguel P, Tobe K, Koyasu S, Taira K, Kitamura T, Shimizu T, Nagai R, Kadowaki T (2003). "Cloning of adiponectin receptors that mediate antidiabetic metabolic effects". Nature. 423 (6941): 762–9. doi:10.1038/nature01705. PMID 12802337.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Grigorieff N, Ceska TA, Downing KH, Baldwin JM, Henderson R (1996). "Electron-crystallographic refinement of the structure of bacteriorhodopsin". J. Mol. Biol. 259 (3): 393–421. doi:10.1006/jmbi.1996.0328. PMID 8676377.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Kimura Y, Vassylyev DG, Miyazawa A, Kidera A, Matsushima M, Mitsuoka K, Murata K, Hirai T, Fujiyoshi Y (1997). "Surface of bacteriorhodopsin revealed by high-resolution electron crystallography". Nature. 389 (6647): 206–11. doi:10.1038/38323. PMID 9296502.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Pebay-Peyroula E, Rummel G, Rosenbusch JP, Landau EM (1997). "X-ray structure of bacteriorhodopsin at 2.5 angstroms from microcrystals grown in lipidic cubic phases". Science. 277 (5332): 1676–81. doi:10.1126/science.277.5332.1676. PMID 9287223.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Trong IL, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M (2000). "Crystal structure of rhodopsin: A G protein-coupled receptor". Science. 289 (5480): 739–45. doi:10.1126/science.289.5480.739. PMID 10926528.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Rasmussen SG, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VR, Sanishvili R, Fischetti RF, Schertler GF, Weis WI, Kobilka BK (2007). "Crystal structure of the human β2-adrenergic G-protein-coupled receptor". Nature. 450 (7168): 383–7. doi:10.1038/nature06325. PMID 17952055.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC (2007). "High-resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor". Science. 318 (5854): 1258–65. doi:10.1126/science.1150577. PMC 2583103. PMID 17962520.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Yao XJ, Weis WI, Stevens RC, Kobilka BK (2007). "GPCR engineering yields high-resolution structural insights into β2-adrenergic receptor function". Science. 318 (5854): 1266–73. doi:10.1126/science.1150609. PMID 17962519.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Rasmussen SG, Choi HJ, Fung JJ, Pardon E, Casarosa P, Chae PS, Devree BT, Rosenbaum DM, Thian FS, Kobilka TS, Schnapp A, Konetzki I, Sunahara RK, Gellman SH, Pautsch A, Steyaert J, Weis WI, Kobilka BK (2011). "Structure of a nanobody-stabilized active state of the β(2) adrenoceptor". Nature. 469 (7329): 175–80. doi:10.1038/nature09648. PMC 3058308. PMID 21228869.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Rosenbaum DM, Zhang C, Lyons JA, Holl R, Aragao D, Arlow DH, Rasmussen SG, Choi HJ, Devree BT, Sunahara RK, Chae PS, Gellman SH, Dror RO, Shaw DE, Weis WI, Caffrey M, Gmeiner P, Kobilka BK (2011). "Structure and function of an irreversible agonist-β(2) adrenoceptor complex". Nature. 469 (7329): 236–40. doi:10.1038/nature09665. PMC 3074335. PMID 21228876.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Warne T, Moukhametzianov R, Baker JG, Nehmé R, Edwards PC, Leslie AG, Schertler GF, Tate CG (2011). "The structural basis for agonist and partial agonist action on a β(1)-adrenergic receptor". Nature. 469 (7329): 241–4. doi:10.1038/nature09746. PMC 3023143. PMID 21228877.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Xu F, Wu H, Katritch V, Han GW, Jacobson KA, Gao ZG, Cherezov V, Stevens RC (2011). "Structure of an agonist-bound human A2A adenosine receptor". Science. 332 (6027): 322–7. doi:10.1126/science.1202793. PMC 3086811. PMID 21393508.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Rasmussen SG, Devree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, Mathiesen JM, Shah ST, Lyons JA, Caffrey M, Gellman SH, Steyaert J, Skiniotis G, Weis WI, Sunahara RK, Kobilka BK (2011). "Crystal structure of the β(2) adrenergic receptor-Gs protein complex". Nature. 477 (7366): 549–55. doi:10.1038/nature10361. PMC 3184188. PMID 21772288.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Lohse MJ, Benovic JL, Codina J, Caron MG, Lefkowitz RJ (1990). "β-Arrestin: a protein that regulates β-adrenergic receptor function". Science. 248 (4962): 1547–1550. doi:10.1126/science.2163110. PMID 2163110.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Luttrell LM, Lefkowitz RJ (2002). "The role of beta-arrestins in the termination and transduction of G-protein-coupled receptor signals". J. Cell. Sci. 115 (Pt 3): 455–65. PMID 11861753.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Millar RP, Newton CL (2010). "The year in G protein-coupled receptor research". Mol. Endocrinol. 24 (1): 261–74. doi:10.1210/me.2009-0473. PMID 20019124.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Brass LF (2003). "Thrombin and platelet activation". Chest. 124 (3 Suppl): 18S–25S. doi:10.1378/chest.124.3_suppl.18S. PMID 12970120.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Rubenstein, Lester A. and Lanzara, Richard G. (1998). "Activation of G protein-coupled receptors entails cysteine modulation of agonist binding". Journal of Molecular Structure (Theochem). 430: 57–71. doi:10.1016/S0166-1280(98)90217-2.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ http://www.bio-balance.com/
- ^ Kim JY, Haastert PV, Devreotes PN (1996). "Social senses: G-protein-coupled receptor signaling pathways in Dictyostelium discoideum". Chem. Biol. 3 (4): 239–43. doi:10.1016/S1074-5521(96)90103-9. PMID 8807851.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Duchene J, Schanstra JP, Pecher C, Pizard A, Susini C, Esteve JP, Bascands JL, Girolami JP (2002). "A novel protein-protein interaction between a G protein-coupled receptor and the phosphatase SHP-2 is involved in bradykinin-induced inhibition of cell proliferation". J Biol Chem. 277 (43): 40375–83. doi:10.1074/jbc.M202744200. PMID 12177051.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Chen-Izu Y, Xiao RP, Izu LT, Cheng H, Kuschel M, Spurgeon H, Lakatta EG (2000). "G(i)-dependent localization of beta(2)-adrenergic receptor signaling to L-type Ca(2+) channels". Biophys. J. 79 (5): 2547–56. doi:10.1016/S0006-3495(00)76495-2. PMC 1301137. PMID 11053129.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ a b Tan CM, Brady AE, Nickols HH, Wang Q, Limbird LE (2004). "Membrane trafficking of G protein-coupled receptors". Annu. Rev. Pharmacol. Toxicol. 44: 559–609. doi:10.1146/annurev.pharmtox.44.101802.121558. PMID 14744258.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Krueger KM, Daaka Y, Pitcher JA, Lefkowitz RJ (1997). "The role of sequestration in G protein-coupled receptor resensitization. Regulation of β2-adrenergic receptor dephosphorylation by vesicular acidification". J. Biol. Chem. 272 (1): 5–8. doi:10.1074/jbc.272.1.5. PMID 8995214.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Laporte SA, Oakley RH, Holt JA, Barak LS, Caron MG (2000). "The interaction of β-arrestin with the AP-2 adaptor is required for the clustering of β2-adrenergic receptor into clathrin-coated pits". J. Biol. Chem. 275 (30): 23120–6. doi:10.1074/jbc.M002581200. PMID 10770944.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Laporte SA, Oakley RH, Zhang J, Holt JA, Ferguson SS, Caron MG, Barak LS (1999). "The beta2-adrenergic receptor/betaarrestin complex recruits the clathrin adaptor AP-2 during endocytosis". Proc. Natl. Acad. Sci. U.S.A. 96 (7): 3712–7. doi:10.1073/pnas.96.7.3712. PMC 22359. PMID 10097102.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Margeta-Mitrovic M, Jan YN, Jan LY (2000). "A trafficking checkpoint controls GABA(B) receptor heterodimerization". Neuron. 27 (1): 97–106. doi:10.1016/S0896-6273(00)00012-X. PMID 10939334.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ White JH, Wise A, Main MJ, Green A, Fraser NJ, Disney GH, Barnes AA, Emson P, Foord SM, Marshall FH (1998). "Heterodimerization is required for the formation of a functional GABA(B) receptor". Nature. 396 (6712): 679–82. doi:10.1038/25354. PMID 9872316.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Krishnan A, Alme´n MS, Fredriksson R, Schiöth HB (2012). "The Origin of GPCRs: Identification of Mammalian like Rhodopsin, Adhesion, Glutamate and Frizzled GPCRs in Fungi". PLoS ONE. 7 (1): e29817. doi:10.1371/journal.pone.0029817. PMC 3251606. PMID 22238661.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Nordström KJ, Sällman Almén M, Edstam MM, Fredriksson R, Schiöth HB (2011). "Independent HHsearch, Needleman–Wunsch-Based, and Motif Analyses Reveal the Overall Hierarchy for Most of the G Protein-Coupled Receptor Families". Mol Biol Evol. 28 (9): 2471–80. doi:10.1093/molbev/msr061. PMID 21402729.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Bakthavatsalam D, Brazill D, Gomer RH, Eichinger L, Rivero F, Noegel AA (2007). "A G protein-coupled receptor with a lipid kinase domain is involved in cell-density sensing". Curr Biol. 17 (10): 892–7. doi:10.1016/j.cub.2007.04.029. PMID 17481898.
{{cite journal}}: CS1 maint: multiple names: authors list (link)
External links
- G-protein-coupled+receptors at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- Wikipedia:MeSH D12.776#MeSH D12.776.543.750.100 --- receptors.2C g-protein-coupled
- "GPCR Database". IUPHAR Database. International Union of Basic and Clinical Pharmacology. Retrieved 2008-08-11.
{{cite web}}: Cite has empty unknown parameter:|coauthors=(help) - Vriend G, Horn F (2006-06-29). "GPCRDB: Information system for G protein-coupled receptors (GPCRs)". Molecular Class-Specific Information System (MCSIS) project. Retrieved 2008-08-11.
{{cite web}}: Cite has empty unknown parameter:|coauthors=(help) - "G Protein-Coupled Receptors on the NET". Retrieved 2010-11-10.
a classification of GPCRs
{{cite web}}: Cite has empty unknown parameter:|coauthors=(help)
Further reading
- "A phylogenetic tree of all human GPCRs" (PDF). Vassilatis DK, Hohmann JG, Zeng H, Li F, Ranchalis JE, Mortrud MT, Brown A, Rodriguez SS, Weller JR, Wright AC, Bergmann JE, Gaitanaris GA (2003). "The G protein-coupled receptor repertoires of human and mouse". Proc Natl Acad Sci USA. 100 (8): 4903–8. doi:10.1073/pnas.0230374100. PMC 153653. PMID 12679517.
{{cite journal}}: CS1 maint: multiple names: authors list (link). Retrieved 2008-08-11.{{cite web}}: Cite has empty unknown parameter:|coauthors=(help); External link in|work=|work=(help); templatestyles stripmarker in|work=at position 1 (help) - "GPCR Reference Library". Retrieved 2008-08-11.
Reference for molecular and mathematical models for the initial receptor response
{{cite web}}: Cite has empty unknown parameter:|coauthors=(help)
