TBX3: Difference between revisions
No edit summary |
No edit summary |
||
| Line 19: | Line 19: | ||
== Role in stem cells == |
== Role in stem cells == |
||
Embryonic stem cells (ESCs) and adult stem cells, are undifferentiated cells which when they divide have the potential to either remain a stem cell or to differentiate into other |
[[Embryonic stem cell|Embryonic stem cells]] (ESCs) and adult stem cells, are undifferentiated cells which when they divide have the potential to either remain a stem cell or to differentiate into other specialized cells. Adult stem cells are multipotent progenitor cells found in numerous adult tissues and, as part of the body repair system, they can develop into more than one cell type but they are more limited than ESCs. <ref>{{Cite journal|last=Gilbert|first=Penney M.|last2=Corbel|first2=Stephane|last3=Doyonnas|first3=Regis|last4=Havenstrite|first4=Karen|last5=Magnusson|first5=Klas E. G.|last6=Blau|first6=Helen M.|date=2012-02-10|title=A single cell bioengineering approach to elucidate mechanisms of adult stem cell self-renewal|url=http://dx.doi.org/10.1039/c2ib00148a|journal=Integrative Biology|volume=4|issue=4|pages=360–367|doi=10.1039/c2ib00148a|issn=1757-9708}}</ref> TBX3 is highly expressed in mouse ESCs (mESCs) and appears to have a dual role in these cells. Firstly it can enhance and maintain stem cell pluripotency by preventing differentiation and enhancing self-renewal and secondly it can maintain the pluripotency and differentiation potential of mESCS. <ref>{{Cite journal|last=Lu|first=Rui|last2=Yang|first2=Acong|last3=Jin|first3=Ying|date=2010-12-28|title=Dual Functions of T-Box 3 (Tbx3) in the Control of Self-renewal and Extraembryonic Endoderm Differentiation in Mouse Embryonic Stem Cells|url=http://dx.doi.org/10.1074/jbc.m110.202150|journal=Journal of Biological Chemistry|volume=286|issue=10|pages=8425–8436|doi=10.1074/jbc.m110.202150|issn=0021-9258}}</ref><ref>{{Cite journal|last=Russell|first=Ronan|last2=Ilg|first2=Marcus|last3=Lin|first3=Qiong|last4=Wu|first4=Guangming|last5=Lechel|first5=André|last6=Bergmann|first6=Wendy|last7=Eiseler|first7=Tim|last8=Linta|first8=Leonhard|last9=Kumar P.|first9=Pavan|date=2015-12|title=A Dynamic Role of TBX3 in the Pluripotency Circuitry|url=http://dx.doi.org/10.1016/j.stemcr.2015.11.003|journal=Stem Cell Reports|volume=5|issue=6|pages=1155–1170|doi=10.1016/j.stemcr.2015.11.003|issn=2213-6711}}</ref> Induced pluripotent stem cells (iPSCs) are ESC-like cells that can generate scalable quantities of relevant tissue and are of major interest for their application in personalized regenerative medicine, drug screening, and for our understanding of the cell signaling networks that regulate embryonic development and disease. In vitro studies have shown that Tbx3 is an important factor that, together with KLF4, SOX2, OCT4, Nanog, LIN-28A and C-MYC, can reprogram somatic cells to form iPS cells. <ref>{{Cite journal|last=Han|first=Jianyong|last2=Yuan|first2=Ping|last3=Yang|first3=Henry|last4=Zhang|first4=Jinqiu|last5=Soh|first5=Boon Seng|last6=Li|first6=Pin|last7=Lim|first7=Siew Lan|last8=Cao|first8=Suying|last9=Tay|first9=Junliang|date=2010-02|title=Tbx3 improves the germ-line competency of induced pluripotent stem cells|url=http://dx.doi.org/10.1038/nature08735|journal=Nature|volume=463|issue=7284|pages=1096–1100|doi=10.1038/nature08735|issn=0028-0836}}</ref> |
||
== Clinical significance == |
== Clinical significance == |
||
TBX3 has been implicated in human diseases including the ulnar mammary syndrome <ref>{{Cite journal|last=Frank|first=Deborah U.|last2=Emechebe|first2=Uchenna|last3=Thomas|first3=Kirk R.|last4=Moon|first4=Anne M.|date=2013-07-02|editor-last=Dettman|editor-first=Robert|title=Mouse Tbx3 Mutants Suggest Novel Molecular Mechanisms for Ulnar-Mammary Syndrome|url=https://dx.plos.org/10.1371/journal.pone.0067841|journal=PLoS ONE|language=en|volume=8|issue=7|pages=e67841|doi=10.1371/journal.pone.0067841|issn=1932-6203|pmc=PMC3699485|pmid=23844108}}</ref>, obesity <ref>{{Cite journal|last=Quarta|first=Carmelo|last2=Fisette|first2=Alexandre|last3=Xu|first3=Yanjun|last4=Colldén|first4=Gustav|last5=Legutko|first5=Beata|last6=Tseng|first6=Yu-Ting|last7=Reim|first7=Alexander|last8=Wierer|first8=Michael|last9=De Rosa|first9=Maria Caterina|date=2019-01-28|title=Functional identity of hypothalamic melanocortin neurons depends on Tbx3|url=http://dx.doi.org/10.1038/s42255-018-0028-1|journal=Nature Metabolism|volume=1|issue=2|pages=222–235|doi=10.1038/s42255-018-0028-1|issn=2522-5812}}</ref>, rheumatoid arthritis <ref>{{Cite journal|last=Sardar|first=Samra|last2=Kerr|first2=Alish|last3=Vaartjes|first3=Daniëlle|last4=Moltved|first4=Emilie Riis|last5=Karosiene|first5=Edita|last6=Gupta|first6=Ramneek|last7=Andersson|first7=Åsa|date=2019-01-10|title=The oncoprotein TBX3 is controlling severity in experimental arthritis|url=http://dx.doi.org/10.1186/s13075-018-1797-3|journal=Arthritis Research & Therapy|volume=21|issue=1|doi=10.1186/s13075-018-1797-3|issn=1478-6362}}</ref> and cancer. <ref>{{Cite journal|last=Willmer|first=Tarryn|last2=Cooper|first2=Aretha|last3=Peres|first3=Jade|last4=Omar|first4=Rehana|last5=Prince|first5=Sharon|date=2017|title=The T-Box transcription factor 3 in development and cancer|url=https://www.jstage.jst.go.jp/article/bst/11/3/11_2017.01043/_article|journal=BioScience Trends|language=en|volume=11|issue=3|pages=254–266|doi=10.5582/bst.2017.01043|issn=1881-7815}}</ref> |
|||
TBX3 has been implicated in human diseases including the ulnar mammary syndrome, obesity, rheumatoid arthritis and cancer <sup>42,67–69</sup>. |
|||
In humans, heterozygous mutations of TBX3 lead to the autosomal dominant developmental disorder, ulnar mammary syndrome (UMS), which is characterized by a number of clinical features including mammary and apocrine gland hypoplasia, upper limb defects, malformations of areola, dental structures, heart and genitalia <sup>2,70</sup>. Several UMS causing mutations in the TBX3 gene have been reported which include 5 nonsense, 8 frameshift (due to deletion, duplication and insertion), 3 missense and 2 splice site mutations. Missense mutations within the T-domain, or the loss of RD1 result in aberrant transcripts and truncated proteins of TBX3. These mutations lead to reduced DNA binding, transcriptional control and splicing regulation of TBX3 and the loss of function and are associated with the most severe phenotype of UMS <sup>16,71–73</sup>. |
In humans, heterozygous mutations of TBX3 lead to the autosomal dominant developmental disorder, ulnar mammary syndrome (UMS), which is characterized by a number of clinical features including mammary and apocrine gland hypoplasia, upper limb defects, malformations of areola, dental structures, heart and genitalia <sup>2,70</sup>. Several UMS causing mutations in the TBX3 gene have been reported which include 5 nonsense, 8 frameshift (due to deletion, duplication and insertion), 3 missense and 2 splice site mutations. Missense mutations within the T-domain, or the loss of RD1 result in aberrant transcripts and truncated proteins of TBX3. These mutations lead to reduced DNA binding, transcriptional control and splicing regulation of TBX3 and the loss of function and are associated with the most severe phenotype of UMS <sup>16,71–73</sup>. |
||
Revision as of 18:17, 30 July 2019
T-box transcription factor TBX3 is a protein that in humans is encoded by the TBX3 gene.[1][2]
T-box 3 (TBX3) is a member of the T-box gene family of transcription factors which all share a highly conserved DNA binding domain known as the T-box. The T-box gene family consists of 17 members in mouse and humans that are grouped into five subfamilies, namely Brachyury (T), T-brain (Tbr1), TBX1, TBX2, and TBX6. Tbx3 is a member of the Tbx2 subfamily which includes Tbx2, Tbx4 and Tbx5.[3] The human TBX3 gene maps to chromosome 12 at position 12q23-24.1 and consists of 7 exons which encodes a 723 amino acid protein (ENSEMBL assembly release GRCh38.p12).
Transcript splicing
Alternative processing and splicing results in at least 4 distinct TBX3 isoforms with TBX3 and TBX3+2a being the predominant isoforms. TBX3+2a results from alternative splicing of the second intron which leads to the addition of the +2a exon and consequently this isoform has an additional 20 amino acids within the T-box DNA binding domain.[8][9] The functions of TBX3 and TBX3+2a may vary slightly across different cell types. [9][10][11][12][13][14]
Structure and function
TBX3 has domains which are important for its transcription factor function which include a DNA-binding domain (DBD) also called the T-box, a nuclear localization signal, two repression domains (R2 and R1) and an activation domain (A).[15] The T-box recognizes a palindromic DNA sequence (T(G/C)ACACCT AGGTGTGAAATT) known as the T-element, or half sites within this sequence called half T-elements, although it can also recognize variations within the consensus T-element sequences. While there are 29 predicted phosphorylation sites in the TBX3 protein only the SP190, SP692 and S720 have been fully characterized. The kinases involved are cyclin A-CDK2 at either SP190 or SP354, p38 mitogen-activated protein (MAP) kinase at SP692 in embryonic kidney cells and AKT3 at S720 in melanoma. These modifications act in a context dependent manner to promote TBX3 protein stability, nuclear localization and transcriptional activity.[16][17][18]
TBX3 can activate and/or repress its target genes by binding a T-element, or half T-element sites.[19] Indeed, Tbx3 binds highly conserved T-elements to activate the promoters of Eomes, T, Sox17 and Gata6, which are factors essential for mesoderm differentiation and extra embryonic endodermal.[20][21] Furthermore, in the cancer context, TBX3 directly represses the cell cycle regulators p19ARF/p14ARF, p21WAF1 and TBX2 as well as E-cadherin which encodes a cell adhesion molecule, to promote proliferation and migration.[11][22][23][24] TBX3 directly represses a region of the PTEN promoter which lacks putative T-elements, but which forms an important regulatory unit for PTEN transcriptional activators, thus raising the possibility that TBX3 may also repress some of its target genes through interfering with transcriptional activators.[25]
The function of TBX3 as either a transcriptional repressor or transcriptional activator is, in part, modulated by protein co-factors. For example, it can interact with other transcription factors such as Nkx2-5, Msx 1/2 [26] and Sox4 [27] to assist it binding to its target genes to regulate heart development [10][28][29][30][31] and it can interact with histone deacetylases (HDACs) 1, 2, 3 and 5 to repress p14ARF in breast cancer and with HDAC5 to repress E-cadherin to promote metastasis in hepatocellular carcinoma. [32][33] Lastly, TBX3 can also co-operate with other factors to inhibit the process of mRNA splicing by directly binding RNAs containing the core motif of a T-element. [10][11][12][13][14] Indeed, TBX3 interacts with Coactivator of AP1 and Estrogen Receptor (CAPERα) to repress the long non-coding RNA, Urothelial Cancer Associated 1 (UCA1), which leads to the bypass of senescence through the stabilization of p16INK4a mRNA. [34]
Role in development
During mouse embryonic development, Tbx3 is expressed in the inner cell mass of the blastocyst, in the extraembryonic mesoderm during gastrulation, and in the developing heart, limbs [35], musculoskeletal structures [36], mammary glands [37], nervous system [38], skin [39], eye [40], liver [41], pancreas [42], lungs [43] and genitalia. [8] Tbx3 null embryos show defects in, among other structures, the heart, mammary glands and limbs and they die in utero by embryonic day E16.5, most likely due to yolk sac and heart defects. These observations together with numerous other studies have illustrated that Tbx3 plays crucial roles in the development of the heart [44], mammary glands [45], limbs [46] and lungs. [47]
Role in stem cells
Embryonic stem cells (ESCs) and adult stem cells, are undifferentiated cells which when they divide have the potential to either remain a stem cell or to differentiate into other specialized cells. Adult stem cells are multipotent progenitor cells found in numerous adult tissues and, as part of the body repair system, they can develop into more than one cell type but they are more limited than ESCs. [48] TBX3 is highly expressed in mouse ESCs (mESCs) and appears to have a dual role in these cells. Firstly it can enhance and maintain stem cell pluripotency by preventing differentiation and enhancing self-renewal and secondly it can maintain the pluripotency and differentiation potential of mESCS. [49][50] Induced pluripotent stem cells (iPSCs) are ESC-like cells that can generate scalable quantities of relevant tissue and are of major interest for their application in personalized regenerative medicine, drug screening, and for our understanding of the cell signaling networks that regulate embryonic development and disease. In vitro studies have shown that Tbx3 is an important factor that, together with KLF4, SOX2, OCT4, Nanog, LIN-28A and C-MYC, can reprogram somatic cells to form iPS cells. [51]
Clinical significance
TBX3 has been implicated in human diseases including the ulnar mammary syndrome [52], obesity [53], rheumatoid arthritis [54] and cancer. [55]
In humans, heterozygous mutations of TBX3 lead to the autosomal dominant developmental disorder, ulnar mammary syndrome (UMS), which is characterized by a number of clinical features including mammary and apocrine gland hypoplasia, upper limb defects, malformations of areola, dental structures, heart and genitalia 2,70. Several UMS causing mutations in the TBX3 gene have been reported which include 5 nonsense, 8 frameshift (due to deletion, duplication and insertion), 3 missense and 2 splice site mutations. Missense mutations within the T-domain, or the loss of RD1 result in aberrant transcripts and truncated proteins of TBX3. These mutations lead to reduced DNA binding, transcriptional control and splicing regulation of TBX3 and the loss of function and are associated with the most severe phenotype of UMS 16,71–73.
Tbx3 is expressed in heterogenous populations of hypothalamic arcuate nucleus neurons which control energy homeostasis by regulating appetite and energy expenditure and the ablation of TBX3 function in these neurons was shown to cause obesity in mouse models. Importantly, Tbx3 was shown to be a key player in driving the functional heterogeneity of hypothalamic neurons and this function was conserved in mice, drosophila and humans 42. Genome wide association studies also casually linked TBX3 to rheumatoid arthritis (RA) susceptibility and a recent study identified Tbx3 as a candidate gene for RA in collagen-induced arthritis (CIA) mouse models 68,74. The severity of RA directly correlated with TBX3 serum levels in the CIA mouse models. Furthermore, Tbx3 was shown to repress B lymphocyte proliferation and to activate the humoral immune response which is associated with chronic inflammation of the synovium leading to RA. Tbx3 may thus be an important player in regulating the immune system and could be used as a biomarker for the diagnosis of RA severity 68.
TBX3 is overexpressed in a wide range of carcinomas (breast, pancreatic, melanoma, liver, lung, gastric, ovarian, bladder and head and neck cancers) and sarcomas (chondrosarcoma, fibrosarcoma, liposarcoma, rhabdomyosarcoma and synovial sarcoma) and there is compelling evidence that it contributes to several hallmarks of cancer. Indeed, TBX3 can bypass cellular senescence, apoptosis and anoikis as well as promote uncontrolled cell proliferation, tumour formation, angiogenesis and metastasis 8,27,69,75–77. Furthermore, TBX3 contributes to the expansion of cancer stem cells (CSCs) and is a key player in regulating pluripotency-related genes in these cells. CSCs contribute to tumour relapse and drug resistance and thus this may be another mechanism by which TBX3 contributes to cancer formation and tumour aggressiveness 96. The mechanisms by which TBX3 contributes to oncogenic processes involve, in part, its ability to inhibit the tumour suppressor pathways p14ARF/p53/p21WAF1/CIP1, p16INK4a/pRb, p57KIP2, PTEN and E-cadherin 9,19,26,78–82 and activating the angiogenesis-associated genes FGF2 and VEGF-A and the EMT gene SNAI 7,8,83. Some of the oncogenic signalling molecules identified that upregulate TBX3 include TGF-β, BRAF-MAPK, c-Myc, AKT and PLCᗴ/PKC 12,18,84–87 The function of TBX3 is also regulated by phosphorylation by the p38-MAPK, AKT3 and cyclin A/CDK2 10 and by protein co-factors, which include PRC2, Histone Deacetylases 1, 2, 3 and 5 and CAPERα 26,28,78.
There is also evidence that TBX3 may function as a tumour suppressor. During oncogenesis, TBX3 is silenced by methylation in some cancers and this was associated with a poor overall survival, resistance to cancer therapy and a more invasive phenotype 88–97. In addition, TBX3 is overexpressed in fibrosarcoma cells and removing TBX3 from these cells led to a more aggressive phenotype 98.
References
- ^ Li QY, Newbury-Ecob RA, Terrett JA, Wilson DI, Curtis AR, Yi CH, Gebuhr T, Bullen PJ, Robson SC, Strachan T, Bonnet D, Lyonnet S, Young ID, Raeburn JA, Buckler AJ, Law DJ, Brook JD (Jan 1997). "Holt-Oram syndrome is caused by mutations in TBX5, a member of the Brachyury (T) gene family". Nature Genetics. 15 (1): 21–9. doi:10.1038/ng0197-21. PMID 8988164.
- ^ "Entrez Gene: TBX3 T-box 3 (ulnar mammary syndrome)".
- ^ Papaioannou VE (October 2014). "The T-box gene family: emerging roles in development, stem cells and cancer". Development. 141 (20): 3819–33. doi:10.1242/dev.104471. PMC 4197708. PMID 25294936.
- ^ a b c GRCh38: Ensembl release 89: ENSG00000135111 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000018604 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ a b Bamshad M, Lin RC, Law DJ, Watkins WC, Krakowiak PA, Moore ME, et al. (July 1997). "Mutations in human TBX3 alter limb, apocrine and genital development in ulnar-mammary syndrome". Nature Genetics. 16 (3): 311–5. doi:10.1038/ng0797-311. PMID 9207801.
- ^ a b Fan W, Huang X, Chen C, Gray J, Huang T (August 2004). "TBX3 and its isoform TBX3+2a are functionally distinctive in inhibition of senescence and are overexpressed in a subset of breast cancer cell lines". Cancer Research. 64 (15): 5132–9. doi:10.1158/0008-5472.CAN-04-0615. PMID 15289316.
- ^ a b c Hoogaars WM, Barnett P, Rodriguez M, Clout DE, Moorman AF, Goding CR, Christoffels VM (June 2008). "TBX3 and its splice variant TBX3 + exon 2a are functionally similar". Pigment Cell & Melanoma Research. 21 (3): 379–87. doi:10.1111/j.1755-148X.2008.00461.x. PMID 18444963.
- ^ a b c Rodriguez M, Aladowicz E, Lanfrancone L, Goding CR (October 2008). "Tbx3 represses E-cadherin expression and enhances melanoma invasiveness". Cancer Research. 68 (19): 7872–81. doi:10.1158/0008-5472.CAN-08-0301. PMID 18829543.
- ^ a b Zhao D, Wu Y, Chen K (February 2014). "Tbx3 isoforms are involved in pluripotency maintaining through distinct regulation of Nanog transcriptional activity". Biochemical and Biophysical Research Communications. 444 (3): 411–4. doi:10.1016/j.bbrc.2014.01.093. PMID 24472544.
- ^ a b Krstic M, Macmillan CD, Leong HS, Clifford AG, Souter LH, Dales DW, et al. (August 2016). "The transcriptional regulator TBX3 promotes progression from non-invasive to invasive breast cancer". BMC Cancer. 16 (1): 671. doi:10.1186/s12885-016-2697-z. PMC 4994202. PMID 27553211.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b Krstic M, Kolendowski B, Cecchini MJ, Postenka CO, Hassan HM, Andrews J, et al. (June 2019). "TBX3 promotes progression of pre-invasive breast cancer cells by inducing EMT and directly up-regulating SLUG". The Journal of Pathology. 248 (2): 191–203. doi:10.1002/path.5245. PMC 6593675. PMID 30697731.
- ^ Carlson H, Ota S, Song Y, Chen Y, Hurlin PJ (May 2002). "Tbx3 impinges on the p53 pathway to suppress apoptosis, facilitate cell transformation and block myogenic differentiation". Oncogene. 21 (24): 3827–35. doi:10.1038/sj.onc.1205476. PMID 12032820.
- ^ Willmer T, Peres J, Mowla S, Abrahams A, Prince S (2015-10-02). "The T-Box factor TBX3 is important in S-phase and is regulated by c-Myc and cyclin A-CDK2". Cell Cycle. 14 (19): 3173–83. doi:10.1080/15384101.2015.1080398. PMC 4825571. PMID 26266831.
- ^ Yano T, Yamazaki Y, Adachi M, Okawa K, Fort P, Uji M, et al. (April 2011). "Tara up-regulates E-cadherin transcription by binding to the Trio RhoGEF and inhibiting Rac signaling". The Journal of Cell Biology. 193 (2): 319–32. doi:10.1083/jcb.201009100. PMC 3080255. PMID 21482718.
- ^ Peres J, Mowla S, Prince S (January 2015). "The T-box transcription factor, TBX3, is a key substrate of AKT3 in melanomagenesis". Oncotarget. 6 (3): 1821–33. doi:10.18632/oncotarget.2782. PMC 4359334. PMID 25595898.
- ^ Wilson V, Conlon FL (2002). "The T-box family". Genome Biology. 3 (6): REVIEWS3008. doi:10.1186/gb-2002-3-6-reviews3008. PMC 139375. PMID 12093383.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Weidgang CE, Russell R, Tata PR, Kühl SJ, Illing A, Müller M, et al. (September 2013). "TBX3 Directs Cell-Fate Decision toward Mesendoderm". Stem Cell Reports. 1 (3): 248–65. doi:10.1016/j.stemcr.2013.08.002. PMC 3849240. PMID 24319661.
- ^ Lu R, Yang A, Jin Y (March 2011). "Dual functions of T-box 3 (Tbx3) in the control of self-renewal and extraembryonic endoderm differentiation in mouse embryonic stem cells". The Journal of Biological Chemistry. 286 (10): 8425–36. doi:10.1074/jbc.M110.202150. PMC 3048727. PMID 21189255.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Lingbeek ME, Jacobs JJ, van Lohuizen M (July 2002). "The T-box repressors TBX2 and TBX3 specifically regulate the tumor suppressor gene p14ARF via a variant T-site in the initiator". The Journal of Biological Chemistry. 277 (29): 26120–7. doi:10.1074/jbc.M200403200. PMID 12000749.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Willmer T, Hare S, Peres J, Prince S (March 2016). "The T-box transcription factor TBX3 drives proliferation by direct repression of the p21(WAF1) cyclin-dependent kinase inhibitor". Cell Division. 11 (1): 6. doi:10.1186/s13008-016-0019-0. PMC 4840944. PMID 27110270.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Li J, Ballim D, Rodriguez M, Cui R, Goding CR, Teng H, Prince S (December 2014). "The anti-proliferative function of the TGF-β1 signaling pathway involves the repression of the oncogenic TBX2 by its homologue TBX3". The Journal of Biological Chemistry. 289 (51): 35633–43. doi:10.1074/jbc.M114.596411. PMC 4271245. PMID 25371204.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Burgucu D, Guney K, Sahinturk D, Ozbudak IH, Ozel D, Ozbilim G, Yavuzer U (October 2012). "Tbx3 represses PTEN and is over-expressed in head and neck squamous cell carcinoma". BMC Cancer. 12 (1): 481. doi:10.1186/1471-2407-12-481. PMC 3517435. PMID 23082988.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Boogerd, Kees-Jan; Wong, L.Y. Elaine; Christoffels, Vincent M.; Klarenbeek, Meinke; Ruijter, Jan M.; Moorman, Antoon F.M.; Barnett, Phil (2008-02-19). "Msx1 and Msx2 are functional interacting partners of T-box factors in the regulation of Connexin43". Cardiovascular Research. 78 (3): 485–493. doi:10.1093/cvr/cvn049. ISSN 1755-3245.
- ^ Boogerd, C. J. J.; Wong, L. Y. E.; van den Boogaard, M.; Bakker, M. L.; Tessadori, F.; Bakkers, J.; ‘t Hoen, P. A. C.; Moorman, A. F.; Christoffels, V. M. (2011-05-03). "Sox4 mediates Tbx3 transcriptional regulation of the gap junction protein Cx43". Cellular and Molecular Life Sciences. 68 (23): 3949–3961. doi:10.1007/s00018-011-0693-7. ISSN 1420-682X.
- ^ Bakker, Martijn L.; Boukens, Bastiaan J.; Mommersteeg, Mathilda T.M.; Brons, Janynke F.; Wakker, Vincent; Moorman, Antoon F.M.; Christoffels, Vincent M. (2008-06-06). "Transcription Factor Tbx3 Is Required for the Specification of the Atrioventricular Conduction System". Circulation Research. 102 (11): 1340–1349. doi:10.1161/circresaha.107.169565. ISSN 0009-7330.
- ^ Christoffels, Vincent M.; Habets, Petra E.M.H.; Franco, Diego; Campione, Marina; Jong, Frits de; Lamers, Wouter H.; Bao, Zheng-Zheng; Palmer, Steve; Biben, Christine (2000-09). "Chamber Formation and Morphogenesis in the Developing Mammalian Heart". Developmental Biology. 225 (1): 266. doi:10.1006/dbio.2000.9859. ISSN 0012-1606.
{{cite journal}}: Check date values in:|date=(help) - ^ Christoffels, Vincent M.; Hoogaars, Willem M.H.; Tessari, Alessandra; Clout, Danielle E.W.; Moorman, Antoon F.M.; Campione, Marina (2004). "T-box transcription factor Tbx2 represses differentiation and formation of the cardiac chambers". Developmental Dynamics. 229 (4): 763–770. doi:10.1002/dvdy.10487. ISSN 1058-8388.
- ^ Stennard, F. A. (2005-10-12). "T-box transcription factors and their roles in regulatory hierarchies in the developing heart". Development. 132 (22): 4897–4910. doi:10.1242/dev.02099. ISSN 0950-1991.
- ^ Yarosh, W.; Barrientos, T.; Esmailpour, T.; Lin, L.; Carpenter, P. M.; Osann, K.; Anton-Culver, H.; Huang, T. (2008-02-01). "TBX3 Is Overexpressed in Breast Cancer and Represses p14ARF by Interacting with Histone Deacetylases". Cancer Research. 68 (3): 693–699. doi:10.1158/0008-5472.can-07-5012. ISSN 0008-5472.
- ^ Dong, Liang; Lyu, Xiaoming; Faleti, Oluwasijibomi Damola; He, Ming-Liang (2018-09). "The special stemness functions of Tbx3 in stem cells and cancer development". Seminars in Cancer Biology. doi:10.1016/j.semcancer.2018.09.010. ISSN 1044-579X.
{{cite journal}}: Check date values in:|date=(help) - ^ Kumar P, Pavan; Emechebe, Uchenna; Smith, Richard; Franklin, Sarah; Moore, Barry; Yandell, Mark; Lessnick, Stephen L; Moon, Anne M (2014-05-29). "Coordinated control of senescence by lncRNA and a novel T-box3 co-repressor complex". eLife. 3. doi:10.7554/elife.02805. ISSN 2050-084X.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Tümpel, S (2002-10-15). "Regulation of Tbx3 Expression by Anteroposterior Signalling in Vertebrate Limb Development". Developmental Biology. 250 (2): 251–262. doi:10.1016/s0012-1606(02)90762-1. ISSN 0012-1606.
- ^ Colasanto, Mary P.; Eyal, Shai; Mohassel, Payam; Bamshad, Michael; Bonnemann, Carsten G.; Zelzer, Elazar; Moon, Anne M.; Kardon, Gabrielle (2016-12-13). "Development of a subset of forelimb muscles and their attachment sites requires the ulnar-mammary syndrome geneTbx3". Development. 143 (24): e1.1–e1.1. doi:10.1242/dev.147645. ISSN 0950-1991.
- ^ Cho, K.-W.; Kim, J.-Y.; Song, S.-J.; Farrell, E.; Eblaghie, M. C.; Kim, H.-J.; Tickle, C.; Jung, H.-S. (2006-11-07). "Molecular interactions between Tbx3 and Bmp4 and a model for dorsoventral positioning of mammary gland development". Proceedings of the National Academy of Sciences. 103 (45): 16788–16793. doi:10.1073/pnas.0604645103. ISSN 0027-8424. PMC 1636533. PMID 17071745.
{{cite journal}}: CS1 maint: PMC format (link) - ^ Quarta, Carmelo; Fisette, Alexandre; Xu, Yanjun; Colldén, Gustav; Legutko, Beata; Tseng, Yu-Ting; Reim, Alexander; Wierer, Michael; De Rosa, Maria Caterina (2019-01-28). "Functional identity of hypothalamic melanocortin neurons depends on Tbx3". Nature Metabolism. 1 (2): 222–235. doi:10.1038/s42255-018-0028-1. ISSN 2522-5812.
- ^ Ichijo, Ryo; Kobayashi, Hiroki; Yoneda, Saori; Iizuka, Yui; Kubo, Hirokazu; Matsumura, Shigeru; Kitano, Satsuki; Miyachi, Hitoshi; Honda, Tetsuya (2017-09-11). "Tbx3-dependent amplifying stem cell progeny drives interfollicular epidermal expansion during pregnancy and regeneration". Nature Communications. 8 (1). doi:10.1038/s41467-017-00433-7. ISSN 2041-1723.
- ^ Motahari, Zahra; Martinez-De Luna, Reyna I.; Viczian, Andrea S.; Zuber, Michael E. (2016-10-01). "Tbx3 represses bmp4 expression and, with Pax6, is required and sufficient for retina formation". Development. 143 (19): 3560–3572. doi:10.1242/dev.130955. ISSN 0950-1991. PMC 5087613. PMID 27578778.
{{cite journal}}: CS1 maint: PMC format (link) - ^ Suzuki, A.; Sekiya, S.; Buscher, D.; Izpisua Belmonte, J. C.; Taniguchi, H. (2008-03-26). "Tbx3 controls the fate of hepatic progenitor cells in liver development by suppressing p19ARF expression". Development. 135 (9): 1589–1595. doi:10.1242/dev.016634. ISSN 0950-1991.
- ^ Begum, Salma; Papaioannou, Virginia E. (2011-12). "Dynamic expression of Tbx2 and Tbx3 in developing mouse pancreas". Gene Expression Patterns. 11 (8): 476–483. doi:10.1016/j.gep.2011.08.003. ISSN 1567-133X.
{{cite journal}}: Check date values in:|date=(help) - ^ Volckaert, Thomas; De Langhe, Stijn P. (2014-12-29). "Wnt and FGF mediated epithelial-mesenchymal crosstalk during lung development". Developmental Dynamics. 244 (3): 342–366. doi:10.1002/dvdy.24234. ISSN 1058-8388.
- ^ Washkowitz, Andrew J.; Gavrilov, Svetlana; Begum, Salma; Papaioannou, Virginia E. (2012-02-14). "Diverse functional networks of Tbx3 in development and disease". Wiley Interdisciplinary Reviews: Systems Biology and Medicine. 4 (3): 273–283. doi:10.1002/wsbm.1162. ISSN 1939-5094.
{{cite journal}}: line feed character in|title=at position 36 (help) - ^ Rowley, Matthew; Grothey, Erin; Couch, Fergus J. (2004-04). "The Role of Tbx2 and Tbx3 in Mammary Development and Tumorigenesis". Journal of Mammary Gland Biology and Neoplasia. 9 (2): 109–118. doi:10.1023/b:jomg.0000037156.64331.3f. ISSN 1083-3021.
{{cite journal}}: Check date values in:|date=(help) - ^ Sheeba, C.J.; Logan, M.P.O. (2017), "The Roles of T-Box Genes in Vertebrate Limb Development", Current Topics in Developmental Biology, Elsevier, pp. 355–381, ISBN 9780128013809, retrieved 2019-07-30
- ^ Lüdtke, Timo H.; Rudat, Carsten; Wojahn, Irina; Weiss, Anna-Carina; Kleppa, Marc-Jens; Kurz, Jennifer; Farin, Henner F.; Moon, Anne; Christoffels, Vincent M. (2016-10). "Tbx2 and Tbx3 Act Downstream of Shh to Maintain Canonical Wnt Signaling during Branching Morphogenesis of the Murine Lung". Developmental Cell. 39 (2): 239–253. doi:10.1016/j.devcel.2016.08.007. ISSN 1534-5807.
{{cite journal}}: Check date values in:|date=(help); no-break space character in|first7=at position 7 (help); no-break space character in|first9=at position 8 (help); no-break space character in|first=at position 5 (help) - ^ Gilbert, Penney M.; Corbel, Stephane; Doyonnas, Regis; Havenstrite, Karen; Magnusson, Klas E. G.; Blau, Helen M. (2012-02-10). "A single cell bioengineering approach to elucidate mechanisms of adult stem cell self-renewal". Integrative Biology. 4 (4): 360–367. doi:10.1039/c2ib00148a. ISSN 1757-9708.
- ^ Lu, Rui; Yang, Acong; Jin, Ying (2010-12-28). "Dual Functions of T-Box 3 (Tbx3) in the Control of Self-renewal and Extraembryonic Endoderm Differentiation in Mouse Embryonic Stem Cells". Journal of Biological Chemistry. 286 (10): 8425–8436. doi:10.1074/jbc.m110.202150. ISSN 0021-9258.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Russell, Ronan; Ilg, Marcus; Lin, Qiong; Wu, Guangming; Lechel, André; Bergmann, Wendy; Eiseler, Tim; Linta, Leonhard; Kumar P., Pavan (2015-12). "A Dynamic Role of TBX3 in the Pluripotency Circuitry". Stem Cell Reports. 5 (6): 1155–1170. doi:10.1016/j.stemcr.2015.11.003. ISSN 2213-6711.
{{cite journal}}: Check date values in:|date=(help); no-break space character in|last9=at position 6 (help) - ^ Han, Jianyong; Yuan, Ping; Yang, Henry; Zhang, Jinqiu; Soh, Boon Seng; Li, Pin; Lim, Siew Lan; Cao, Suying; Tay, Junliang (2010-02). "Tbx3 improves the germ-line competency of induced pluripotent stem cells". Nature. 463 (7284): 1096–1100. doi:10.1038/nature08735. ISSN 0028-0836.
{{cite journal}}: Check date values in:|date=(help) - ^ Frank, Deborah U.; Emechebe, Uchenna; Thomas, Kirk R.; Moon, Anne M. (2013-07-02). Dettman, Robert (ed.). "Mouse Tbx3 Mutants Suggest Novel Molecular Mechanisms for Ulnar-Mammary Syndrome". PLoS ONE. 8 (7): e67841. doi:10.1371/journal.pone.0067841. ISSN 1932-6203. PMC 3699485. PMID 23844108.
{{cite journal}}: CS1 maint: PMC format (link) CS1 maint: unflagged free DOI (link) - ^ Quarta, Carmelo; Fisette, Alexandre; Xu, Yanjun; Colldén, Gustav; Legutko, Beata; Tseng, Yu-Ting; Reim, Alexander; Wierer, Michael; De Rosa, Maria Caterina (2019-01-28). "Functional identity of hypothalamic melanocortin neurons depends on Tbx3". Nature Metabolism. 1 (2): 222–235. doi:10.1038/s42255-018-0028-1. ISSN 2522-5812.
- ^ Sardar, Samra; Kerr, Alish; Vaartjes, Daniëlle; Moltved, Emilie Riis; Karosiene, Edita; Gupta, Ramneek; Andersson, Åsa (2019-01-10). "The oncoprotein TBX3 is controlling severity in experimental arthritis". Arthritis Research & Therapy. 21 (1). doi:10.1186/s13075-018-1797-3. ISSN 1478-6362.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Willmer, Tarryn; Cooper, Aretha; Peres, Jade; Omar, Rehana; Prince, Sharon (2017). "The T-Box transcription factor 3 in development and cancer". BioScience Trends. 11 (3): 254–266. doi:10.5582/bst.2017.01043. ISSN 1881-7815.
Further reading
- Bamshad M, Lin RC, Law DJ, Watkins WC, Krakowiak PA, Moore ME, et al. (July 1997). "Mutations in human TBX3 alter limb, apocrine and genital development in ulnar-mammary syndrome". Nature Genetics. 16 (3): 311–5. doi:10.1038/ng0797-311. PMID 9207801.
- Bamshad M, Le T, Watkins WS, Dixon ME, Kramer BE, Roeder AD, et al. (June 1999). "The spectrum of mutations in TBX3: Genotype/Phenotype relationship in ulnar-mammary syndrome". American Journal of Human Genetics. 64 (6): 1550–62. doi:10.1086/302417. PMC 1377898. PMID 10330342.
- He M, Wen L, Campbell CE, Wu JY, Rao Y (August 1999). "Transcription repression by Xenopus ET and its human ortholog TBX3, a gene involved in ulnar-mammary syndrome". Proceedings of the National Academy of Sciences of the United States of America. 96 (18): 10212–7. doi:10.1073/pnas.96.18.10212. PMC 17868. PMID 10468588.
- Carlson H, Ota S, Campbell CE, Hurlin PJ (October 2001). "A dominant repression domain in Tbx3 mediates transcriptional repression and cell immortalization: relevance to mutations in Tbx3 that cause ulnar-mammary syndrome". Human Molecular Genetics. 10 (21): 2403–13. doi:10.1093/hmg/10.21.2403. PMID 11689487.
- Brummelkamp TR, Kortlever RM, Lingbeek M, Trettel F, MacDonald ME, van Lohuizen M, Bernards R (February 2002). "TBX-3, the gene mutated in Ulnar-Mammary Syndrome, is a negative regulator of p19ARF and inhibits senescence". The Journal of Biological Chemistry. 277 (8): 6567–72. doi:10.1074/jbc.M110492200. PMID 11748239.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - Lingbeek ME, Jacobs JJ, van Lohuizen M (July 2002). "The T-box repressors TBX2 and TBX3 specifically regulate the tumor suppressor gene p14ARF via a variant T-site in the initiator". The Journal of Biological Chemistry. 277 (29): 26120–7. doi:10.1074/jbc.M200403200. PMID 12000749.
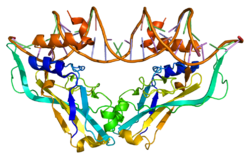
{{cite journal}}: CS1 maint: unflagged free DOI (link) - Coll M, Seidman JG, Müller CW (March 2002). "Structure of the DNA-bound T-box domain of human TBX3, a transcription factor responsible for ulnar-mammary syndrome". Structure. 10 (3): 343–56. doi:10.1016/S0969-2126(02)00722-0. PMID 12005433.
- Carlson H, Ota S, Song Y, Chen Y, Hurlin PJ (May 2002). "Tbx3 impinges on the p53 pathway to suppress apoptosis, facilitate cell transformation and block myogenic differentiation". Oncogene. 21 (24): 3827–35. doi:10.1038/sj.onc.1205476. PMID 12032820.
- Sasaki G, Ogata T, Ishii T, Hasegawa T, Sato S, Matsuo N (July 2002). "Novel mutation of TBX3 in a Japanese family with ulnar-mammary syndrome: implication for impaired sex development". American Journal of Medical Genetics. 110 (4): 365–9. doi:10.1002/ajmg.10447. PMID 12116211.
- Wollnik B, Kayserili H, Uyguner O, Tukel T, Yuksel-Apak M (2003). "Haploinsufficiency of TBX3 causes ulnar-mammary syndrome in a large Turkish family". Annales De Genetique. 45 (4): 213–7. doi:10.1016/S0003-3995(02)01144-9. PMID 12668170.
- Fan W, Huang X, Chen C, Gray J, Huang T (August 2004). "TBX3 and its isoform TBX3+2a are functionally distinctive in inhibition of senescence and are overexpressed in a subset of breast cancer cell lines". Cancer Research. 64 (15): 5132–9. doi:10.1158/0008-5472.CAN-04-0615. PMID 15289316.
- Lomnytska M, Dubrovska A, Hellman U, Volodko N, Souchelnytskyi S (January 2006). "Increased expression of cSHMT, Tbx3 and utrophin in plasma of ovarian and breast cancer patients". International Journal of Cancer. 118 (2): 412–21. doi:10.1002/ijc.21332. PMID 16049973.
- Yang L, Cai CL, Lin L, Qyang Y, Chung C, Monteiro RM, et al. (April 2006). "Isl1Cre reveals a common Bmp pathway in heart and limb development". Development. 133 (8): 1575–85. doi:10.1242/dev.02322. PMC 5576437. PMID 16556916.
- Lee HS, Cho HH, Kim HK, Bae YC, Baik HS, Jung JS (February 2007). "Tbx3, a transcriptional factor, involves in proliferation and osteogenic differentiation of human adipose stromal cells". Molecular and Cellular Biochemistry. 296 (1–2): 129–36. doi:10.1007/s11010-006-9306-4. PMID 16955224.
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M (November 2006). "Global, in vivo, and site-specific phosphorylation dynamics in signaling networks". Cell. 127 (3): 635–48. doi:10.1016/j.cell.2006.09.026. PMID 17081983.
- Mommersteeg MT, Hoogaars WM, Prall OW, de Gier-de Vries C, Wiese C, Clout DE, et al. (February 2007). "Molecular pathway for the localized formation of the sinoatrial node". Circulation Research. 100 (3): 354–62. doi:10.1161/01.RES.0000258019.74591.b3. PMID 17234970.
External links
- TBX3+protein,+human at the U.S. National Library of Medicine Medical Subject Headings (MeSH)