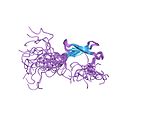
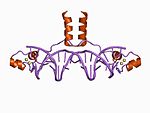
Zinc finger


Zinc fingers are small protein structural motifs that can coordinate one or more zinc ions to help stabilize their folds. They can be classified into several different structural families (zinc finger proteins) and typically function as interaction modules that bind DNA, RNA, proteins, or small molecules. The name "zinc finger" was originally coined to describe the finger-like appearance of a diagram showing the hypothesized structure of the repeated unit in Xenopus laevis transcription factor IIIA.[1]
Classes
Zinc fingers coordinate zinc ions with a combination of cysteine and histidine residues. They can be classified by the type and order of these zinc coordinating residues (e.g., Cys2His2, Cys4, and Cys6). A more systematic method classifies them into different "fold groups" based on the overall shape of the protein backbone in the folded domain. The most common "fold groups" of zinc fingers are the Cys2His2-like (the "classic zinc finger"), treble clef, and zinc ribbon.[2]
The following table[2] shows the different structures and their key features:
Cys2His2
The Cys2His2-like fold group is by far the best-characterized class of zinc fingers and are extremely common in mammalian transcription factors. These domains adopt a simple ββα fold and have the amino acid Sequence motif:[3]
X2-Cys-X2,4-Cys-X12-His-X3,4,5-His
This class of zinc fingers can have a variety of functions such as binding RNA and mediating protein-protein interactions, but is best known for its role in sequence-specific DNA-binding proteins such as Zif268. In such proteins, individual zinc finger domains typically occur as tandem repeats with two, three, or more fingers comprising the DNA-binding domain of the protein. These tandem arrays can bind in the major groove of DNA and are typically spaced at 3-bp intervals. The α-helix of each domain (often called the "recognition helix") can make sequence-specific contacts to DNA bases; residues from a single recognition helix can contact 4 or more bases to yield an overlapping pattern of contacts with adjacent zinc fingers.
Gag-knuckle
| Zinc knuckle | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||
| Symbol | zf-CCHC | ||||||||||
| Pfam | PF00098 | ||||||||||
| InterPro | IPR001878 | ||||||||||
| SMART | SM00343 | ||||||||||
| PROSITE | PS50158 | ||||||||||
| |||||||||||
This fold group is defined by two short β-strands connected by a turn (zinc knuckle) followed by a short helix or loop and resembles the classical Cys2His2 motif with a large portion of the helix and β-hairpin truncated.
The retroviral nucleocapsid (NC) protein from HIV and other related retroviruses are examples of proteins possessing these motifs. The gag-knuckle zinc finger in the HIV NC protein is the target of a class of drugs known as zinc finger inhibitors.
Treble-clef
The treble-clef motif consists of a β-hairpin at the N-terminus and an α-helix at the C-terminus that each contribute two ligands for zinc binding, although a loop and a second β-hairpin of varying length and conformation can be present between the N-terminal β-hairpin and the C-terminal α-helix. These fingers are present in a diverse group of proteins that frequently do not share sequence or functional similarity with each other. The best-characterized proteins containing treble-clef zinc fingers are the nuclear hormone receptors.
Zinc Ribbon
| TFIIB zinc-binding | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||
| Symbol | TF_Zn_Ribbon | ||||||||||
| Pfam | PF08271 | ||||||||||
| Pfam clan | Zn_Beta_Ribbon | ||||||||||
| InterPro | IPR013137 | ||||||||||
| PROSITE | PS51134 | ||||||||||
| |||||||||||
The zinc ribbon fold is characterised by two beta-hairpins forming two structurally similar zinc-binding sub-sites.
Zn2/Cys6
| Fungal Zn(2)-Cys(6) binuclear cluster domain | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||
| Symbol | Zn_clus | ||||||||||
| Pfam | PF00172 | ||||||||||
| InterPro | IPR001138 | ||||||||||
| SMART | GAL4 | ||||||||||
| PROSITE | PS00463 | ||||||||||
| CDD | cd00067 | ||||||||||
| |||||||||||
The canonical members of this class contain a binuclear zinc cluster in which two zinc ions are bound by six cysteine residues. These zinc fingers can be found in several transcription factors including the yeast Gal4 protein.
Engineered zinc finger arrays
Generating arrays of engineered Cys2His2 zinc fingers is the most developed method for creating proteins capable of targeting desired genomic DNA sequences. The majority of engineered zinc finger arrays are based on the zinc finger domain of the murine transcription factor Zif268, although some groups have used zinc finger arrays based on the human transcription factor SP1. Zif268 has three individual zinc finger motifs that collectively bind a 9 bp sequence with high affinity.[4] The structure of this protein bound to DNA was solved in 1991[5] and stimulated a great deal of research into engineered zinc finger arrays. In 1994 and 1995, a number of groups used phage display to alter the specificity of a single zinc finger of Zif268.[6][7][8][9] Carlos F. Barbas et al. also reported the development of zinc finger technology in the patent literature and have been granted a number of patents that have been important for the commercial development of zinc finger technology.[10][11] Typical engineered zinc finger arrays have between 3 and 6 individual zinc finger motifs and bind target sites ranging from 9 basepairs to 18 basepairs in length. Arrays with 6 zinc finger motifs are particularly attractive because they bind a target site that is long enough to have a good chance of being unique in a mammalian genome.[12] There are two main methods currently used to generate engineered zinc finger arrays, modular assembly and a bacterial selection system, and there is some debate about which method is best suited for most applications.[13][14]
Modular assembly
The most straightforward method to generate new zinc finger arrays is to combine smaller zinc finger "modules" of known specificity. The structure of the zinc finger protein Zif268 bound to DNA described by Pavletich and Pabo in their 1991 publication has been key to much of this work and describes the concept of obtaining fingers for each of the 64 possible base pair triplets and then mixing and matching these fingers to design proteins with any desired sequence specificity.[5] The most common modular assembly process involves combining separate zinc fingers that can each recognize a 3 basepair DNA sequence to generate 3-finger, 4-, 5-, or 6-finger arrays that recognize target sites ranging from 9 basepairs to 18 basepairs in length. Another method uses 2-finger modules to generate zinc finger arrays with up to six individual zinc fingers.[15] The Barbas Laboratory of The Scripps Research Institute used phage display to develop and characterize zinc finger domains that recognize most DNA triplet sequences[16][17][18] while another group isolated and characterized individual fingers from the human genome.[19] A potential drawback with modular assembly in general is that specificities of individual zinc finger can overlap and can depend on the context of the surrounding zinc fingers and DNA. A recent study demonstrated that a high proportion of 3-finger zinc finger arrays generated by modular assembly fail to bind their intended target with sufficient affinity in a bacterial two-hybrid assay and fail to function as zinc finger nucleases, but the success rate was somewhat higher when sites of the form GNNGNNGNN were targeted.[20] A subsequent study used modular assembly to generate zinc finger nucleases with both 3-finger arrays and 4-finger arrays and observed a much higher success rate with 4-finger arrays.[21] A variant of modular assembly that takes the context of neighboring fingers into account has also been reported and this method tends to yield proteins with improved performance relative to standard modular assembly.[22]
Selection methods
Numerous selection methods have been used to generate zinc finger arrays capable of targeting desired sequences. Initial selection efforts utilized phage display to select proteins that bound a given DNA target from a large pool of partially randomized zinc finger arrays. This technique is difficult to use on more than a single zinc finger at a time, so a multi-step processes that generated a completely optimized 3-finger array by adding and optimizing a single zinc finger at a time was developed.[23] More recent efforts have utilized yeast one-hybrid systems, bacterial one-hybrid and two-hybrid systems, and mammalian cells. A promising new method to select novel 3-finger zinc finger arrays utilizes a bacterial two-hybrid system and has been dubbed "OPEN" by its creators.[24] This system combines pre-selected pools of individual zinc fingers that were each selected to bind a given triplet and then utilizes a second round of selection to obtain 3-finger arrays capable of binding a desired 9-bp sequence. This system was developed by the Zinc Finger Consortium as an alternative to commercial sources of engineered zinc finger arrays. It is somewhat difficult to directly compare the binding properties of proteins generated with this method to proteins generated by modular assembly as the specificity profiles of proteins generated by the OPEN method have never been reported.
Applications
Engineered zinc finger arrays can then be used in numerous applications such as artificial transcription factors, zinc finger methylases, zinc finger recombinases, and Zinc finger nucleases.[25] While initial studies with another DNA-binding domain from bacterial TAL effectors show promise,[26][27][28][29] it remains to be seen whether these domains are suitable for some or all of the applications where engineered zinc fingers are currently used. Artificial transcription factors with engineered zinc finger arrays have been used in numerous scientific studies, and an artificial transcription factor that activates expression of VEGF is currently being evaluated in humans as a potential treatment for several clinical indications. Zinc finger nucleases have become useful reagents for manipulating genomes of many higher organisms including Drosophila melanogaster, Caenorhabditis elegans, tobacco, corn,[15] zebrafish,[30] various types of mammalian cells,[31] and rats.[32] An ongoing clinical trial is evaluating Zinc finger nucleases that disrupt the CCR5 gene in CD4+ human T-cells as a potential treatment for HIV/AIDS.[33]
See also
- Zinc finger nuclease
- Zinc finger inhibitor
- Steroid hormone receptor
- DNA-binding protein
- Sequence motif
- Structural motif
- RING finger domain
- B-box zinc finger
References
- ^ Miller J, McLachlan AD, Klug A (1985). "Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes". EMBO J. 4 (6): 1609–14. PMC 554390. PMID 4040853.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ a b Krishna SS, Majumdar I, Grishin NV (2003). "Structural classification of zinc fingers: survey and summary". Nucleic Acids Res. 31 (2): 532–50. doi:10.1093/nar/gkg161. PMC 140525. PMID 12527760.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ C.O. Pabo; E.Peisach; R.A. Grant (2001). "Design and Selection of Novel Cys2His2 Zinc Finger Proteins". Annu. Rev. Biochem. 70: 313–40. doi:10.1146/annurev.biochem.70.1.313. PMID 11395410.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^
Christy B, Nathans D (1989). "DNA binding site of the growth factor-inducible protein Zif268". Proc. Natl. Acad. Sci. U.S.A. 86 (22): 8737–41. doi:10.1073/pnas.86.22.8737. PMC 298363. PMID 2510170.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ a b Pavletich NP, Pabo CO (1991). "Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A". Science. 252 (5007): 809–17. doi:10.1126/science.2028256. PMID 2028256.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^
Rebar EJ, Pabo CO (1994). "Zinc finger phage: affinity selection of fingers with new DNA-binding specificities". Science. 263 (5147): 671–3. doi:10.1126/science.8303274. PMID 8303274.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^
Jamieson AC, Kim SH, Wells JA (1994). "In vitro selection of zinc fingers with altered DNA-binding specificity". Biochemistry. 33 (19): 5689–95. doi:10.1021/bi00185a004. PMID 8180194.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^
Choo Y, Klug A (1994). "Toward a code for the interactions of zinc fingers with DNA: selection of randomized fingers displayed on phage". Proc. Natl. Acad. Sci. U.S.A. 91 (23): 11163–7. doi:10.1073/pnas.91.23.11163. PMC 45187. PMID 7972027.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^
Wu H, Yang WP, Barbas CF (1995). "Building zinc fingers by selection: toward a therapeutic application". Proc. Natl. Acad. Sci. U.S.A. 92 (2): 344–8. doi:10.1073/pnas.92.2.344. PMC 42736. PMID 7831288.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ United States Patents 6,140,466, 6,140,081; 6,242,568; 6,610,512; 6,790,941; 7,011,972; 7,067,617; 7,101,972; 7,329,541; 7,151,201; 7,329,728; 7,378,510; 7,442,784; 7,741,110; 7,781,645; 7,833,784; Barbas et al. inventors
- ^ Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1038/nbt0805-915, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1038/nbt0805-915instead. - ^
Liu Q, Segal DJ, Ghiara JB, Barbas CF (1997). "Design of polydactyl zinc-finger proteins for unique addressing within complex genomes". Proc. Natl. Acad. Sci. U.S.A. 94 (11): 5525–30. doi:10.1073/pnas.94.11.5525. PMC 20811. PMID 9159105.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^
Kim JS, Lee HJ, Carroll D (2010). "Genome editing with modularly assembled zinc-finger nucleases". Nat. Methods. 7 (2): 91, author reply 91–2. doi:10.1038/nmeth0210-91a. PMC 2987589. PMID 20111032.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^
Joung JK, Voytas DF, Cathomen T (2010). "Reply to "Genome editing with modularly assembled zinc-finger nucleases"". Nat. Methods. 7 (2): 91–2. doi:10.1038/nmeth0210-91b. PMID 20111031.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ a b Shukla VK, Doyon Y, Miller JC; et al. (2009). "Precise genome modification in the crop species Zea mays using zinc-finger nucleases". Nature. 459 (7245): 437–41. Bibcode:2009Natur.459..437S. doi:10.1038/nature07992. PMID 19404259.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^
Segal DJ, Dreier B, Beerli RR, Barbas CF (1999). "Toward controlling gene expression at will: selection and design of zinc finger domains recognizing each of the 5'-GNN-3' DNA target sequences". Proc. Natl. Acad. Sci. U.S.A. 96 (6): 2758–63. doi:10.1073/pnas.96.6.2758. PMC 15842. PMID 10077584.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^
Dreier B, Fuller RP, Segal DJ; et al. (2005). "Development of zinc finger domains for recognition of the 5'-CNN-3' family DNA sequences and their use in the construction of artificial transcription factors". J. Biol. Chem. 280 (42): 35588–97. doi:10.1074/jbc.M506654200. PMID 16107335.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^
Dreier B, Beerli RR, Segal DJ, Flippin JD, Barbas CF (2001). "Development of zinc finger domains for recognition of the 5'-ANN-3' family of DNA sequences and their use in the construction of artificial transcription factors". J. Biol. Chem. 276 (31): 29466–78. doi:10.1074/jbc.M102604200. PMID 11340073.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^
Bae KH, Kwon YD, Shin HC; et al. (2003). "Human zinc fingers as building blocks in the construction of artificial transcription factors". Nat. Biotechnol. 21 (3): 275–80. doi:10.1038/nbt796. PMID 12592413.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^
C.L. Ramirez; J.E. Foley; D.A. Wright; F. Muller-Lerch; S.H. Rahman; T.I. Cornu; R.J. Winfrey; J.D. Sander; F. Fu; J.A. Townsend; T. Cathomen; D.F. Voytas; J.K. Joung (2009). "Unexpected failure rates for modular assembly of engineered zinc fingers". Nature Methods. 5 (5): 374–375. doi:10.1038/nmeth0508-374. PMID 18446154.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^
Kim HJ, Lee HJ, Kim H, Cho SW, Kim JS (2009). "Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly". Genome Res. 19 (7): 1279–88. doi:10.1101/gr.089417.108. PMC 2704428. PMID 19470664.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1038/nmeth.1542, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1038/nmeth.1542instead. - ^
Greisman HA, Pabo CO (1997). "A general strategy for selecting high-affinity zinc finger proteins for diverse DNA target sites". Science. 275 (5300): 657–61. doi:10.1126/science.275.5300.657. PMID 9005850.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^
M.L. Maeder; et al. (2008). "Rapid "Open-Source" Engineering of Customized Zinc-Finger Nucleases for Highly Efficient Gene Modification". Mol. Cell. 31 (2): 294–301. doi:10.1016/j.molcel.2008.06.016. PMC 2535758. PMID 18657511.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help) - ^
A.C. Jamieson; J.C. Miller; C.O. Pabo (2003). "Drug Discovery with Engineered zinc-finger proteins". Nat. Rev. Drug Discov. 2 (5): 361–8. doi:10.1038/nrd1087. PMID 12750739.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^
Moscou MJ, Bogdanove AJ (2009). "A simple cipher governs DNA recognition by TAL effectors". Science. 326 (5959): 1501. Bibcode:2009Sci...326.1501M. doi:10.1126/science.1178817. PMID 19933106.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Boch J, Scholze H, Schornack S; et al. (2009). "Breaking the code of DNA binding specificity of TAL-type III effectors". Science. 326 (5959): 1509–12. doi:10.1126/science.1178811. PMID 19933107.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Christian M, Cermak T, Doyle EL; et al. (2010). "TAL Effector Nucleases Create Targeted DNA Double-strand Breaks". Genetics. 186 (2): 757–761. doi:10.1534/genetics.110.120717. PMC 2942870. PMID 20660643.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Li T, Huang S, Jiang WZ; et al. (2010). "TAL nucleases (TALNs): hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain". Nucleic Acids Res. 39 (1): 359–372. doi:10.1093/nar/gkq704. PMC 3017587. PMID 20699274.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ S.C. Ekker (2008). "Zinc finger-based knockout punches for zebrafish genes". Zebrafish. 5 (2): 1121–3. doi:10.1089/zeb.2008.9988. PMC 2849655. PMID 18554175.
- ^ D. Carroll (2008). "Progress and prospects: Zinc-finger nucleases as gene therapy agents". Gene Therapy. 15 (22): 1463–1468. doi:10.1038/gt.2008.145. PMC 2747807. PMID 18784746.
- ^
Geurts AM, Cost GJ, Freyvert Y; et al. (2009). "Knockout rats via embryo microinjection of zinc-finger nucleases". Science. 325 (5939): 433. doi:10.1126/science.1172447. PMC 2831805. PMID 19628861.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^
Tebas, Pablo; et al. (2009). "Autologous T-Cells Genetically Modified at the CCR5 Gene by Zinc Finger Nucleases SB-728 for HIV (Zinc-Finger)". ClinicalTrials.gov.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)
External links
- McDowall J. "Protein of the Month: Zinc Fingers". European Molecular Biology Laboratory - European Bioinformatics Institute (EMBL-EBI). Retrieved 2008-01-13.
- Goodsell DS. "Molecule of the Month: Zinc Fingers". Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB). Retrieved 2008-01-13.
- The double helix between the zinc finger
- Zinc Finger Tools design and information site
- Human KZNF Gene Catalog
- Zinc finger C2H2-type domain in PROSITE
- Entry for zinc finger class C2H2 in the SMART database
- The Zinc Finger Consortium
- ZiFiT- Zinc Finger Design Tool
- Zinc Finger Consortium Materials from Addgene