MyoD
Template:PBB MyoD is a protein that plays a major role in regulating muscle differentiation. MyoD, which was discovered in the laboratory of Harold M. Weintraub, belongs to a family of proteins known as myogenic regulatory factors (MRFs).[1] These bHLH (basic helix loop helix) transcription factors act sequentially in myogenic differentiation. MRF family members include MyoD, Myf5, myogenin, and MRF4 (Myf6).
MyoD is one of the earliest markers of myogenic commitment. MyoD is expressed at extremely low and essentially undetectable levels in quiescent satellite cells, but expression of MyoD is activated in response to exercise or muscle tissue damage. The effect of MyoD on satellite cells is dose-dependent; high MyoD expression represses cell renewal and induces apoptosis. Although MyoD marks myoblast commitment, muscle development is not dramatically ablated in mouse mutants lacking the MyoD gene. This is likely due to functional redundancy from Myf5. Nevertheless, the combination of MyoD and Myf5 is vital to the success of myogenesis. MyoD is also an important effector for the fast-twitch muscle fiber (type IIa and IIx) phenotype.[2]
History
MyoD was first described as a nuclear phosphoprotein in 1988 by Tapscott, Davis, Thayer, Cheng, Weintraub, and Lassar in Science. The researchers expressed the complementary DNA (cDNA) of the murine MyoD protein in a different cell lines (fibroblast and adipoblast) and found MyoD converted them to myogenic cells.[3] The following year, the same research team performed several tests to determine both the structure and function of the protein, confirming their initial proposal that the active site of the protein consisted of the helix loop helix (now referred to as basic helix loop helix) for dimerization and a basic site upstream of this bHLH region facilitated DNA binding only once it became a protein dimer.[4] MyoD has since been an active area of research as still relatively little is known concerning many aspects of its function.
Function
The function of MyoD in development is to commit mesoderm cells to a skeletal myoblast lineage, and then to regulate that continued state. MyoD may also play a role in regulating muscle repair. MyoD mRNA levels are also reported to be elevated in aging skeletal muscle.
One of the main actions of MyoD is to remove cells from the cell cycle (halt proliferation for terminal cell cycle arrest in differentiated myoblasts) by enhancing the transcription of p21. MyoD is inhibited by cyclin dependent kinases (CDKs). CDKs are in turn inhibited by p21. Thus MyoD enhances its own activity in the cell in a feedforward manner.
Sustained MyoD expression is necessary for retaining the expression of muscle-related genes.[5]
Mechanisms
MyoD is known to have binding interactions with hundreds of muscular gene promoters and drive myoblast proliferation. While not completely understood, MyoD is now thought to function as the ultimate myogenesis controller in an on/off switch association mediated by KAP1 (KRAB [Krüppel-like associated box]-associated protein 1) phosphorylation.[6] KAP1 is localized at muscle-related genes in myoblasts along with both MyoD and Mef2 (a myocyte transcription enhancer factor). Here, it serves as a scaffold and recruits the coactivators p300 and LSD1, in addition to several corepressors which include G9a and the Histone deacetylase HDAC1. The resulting consequence of the coactivator/corepressor recruitment is silenced promoting regions on muscle genes. When the kinase MSK1 phosphorylates KAP1, the corepressors previously bound to the scaffold are released allowing MyoD and Mef2 to activate transcription.[7]
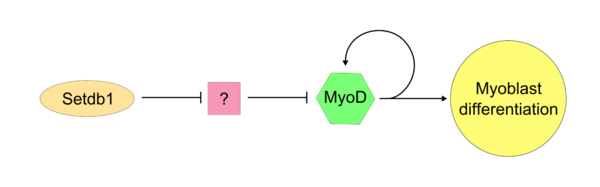
Once the "master controller" MyoD has become active, SETDB1 is required to maintain MyoD expression within the cell. Setdb1 appears to be necessary to maintain both MyoD expression and also genes that are specific to muscle tissues because reduction of Setdb1 expression results in a severe delay of myoblast differentiation and determination.[8] In Setdb1 depleted myoblasts that are treated with exogenous MyoD, myoblastic differentiation is successfully restored. In one model of Setdb1 action on MyoD, Setdb1 represses an inhibitor of MyoD. This unidentified inhibitor likely acts competitively against MyoD during typical cellular proliferation. Evidence for this model is that reduction of Setdb1 results in direct inhibition of myoblast differentiation which may be caused by the release of the unknown MyoD inhibitor.
MyoD has also been shown to function cooperatively with the proto-oncogene, Retinoblastoma (pRb) to cause cell cycle arrest in the terminally differentiated myoblasts.[9] This is done through regulation of the Cyclin, Cyclin D1. Cell cycle arrest (in which myoblasts would indicate the conclusion of myogenesis) is dependent on the continuous and stable repression of the D1 cyclin. Both MyoD and pRb are necessary for the repression of cyclin D1, but rather than acting directly on cyclin D1, they act on Fra-1 which is immediately early of cyclin D1. MyoD and pRb are both necessary for repressing Fra-1 (and thus cyclin D1) as either MyoD or pRb on its own is not sufficient alone to induce cyclin D1 repression and thus cell cycle arrest. In an intronic enhancer of Fra-1 there were two conserved MyoD binding sites discovered. There is cooperative action of MyoD and pRb at the Fra-1 intronic enhancer that suppresses the enhancer, therefore suppressing cyclin D1 and ultimately resulting in cell cycle arrest for terminally differentiated myoblasts.[10]
Role in the wnt signalling pathway
The Wnt signalling pathway family refers to a group of secreted paracrine signaling proteins. Nineteen glycoproteins have been identified as members of this family. Wnts bind to the plasma membrane receptor Frizzled on the target cells. Several different intracellular pathways of signalling are activated by Wnts interacting with the Frizzled receptor. One of the most common is inhibition of glycogen synthase kinase 3β (GSK3β) which functions to destroy β-catenin. This inhibition allows β-catenin to accumulate in the cell cytoplasm and eventual translocation into the nucleus. In the canonical Wnt pathway, nuclear β-catenin forms a dimer with T-cell factor (TCF) and lympoid enhaner factor (LEF) on the gene promoter that is targeted. This initiates transcription and mediates the physiological response to the initial Wnt signal. Wnt-Frizzled pathways also are known to activate c-Jun N-terminal kinases (JNKs) in the cytoplasm. Wnts also drive the translocation of NFATs into the nucleus to initiate the transcription of the targeted genes.[11]
Proteins involved in the Wnt signalling pathway induce cells in somites and competent tissues that receive these wnt signals to actively express Pax3 and Pax7 in addition to myogenic regulatory factors, including Myf5 and MyoD. Specifically, Wnt3a is responsible for direct induction of MyoD expression via cis-element interactions with a distal enhancer and Wnt response element.[12]
In typical adult muscles in a resting condition (absence of physiological stress) the specific Wnt family proteins that are expressed are Wnt5a, Wnt5b, Wnt7a and Wnt4. When a muscle becomes injured (thus requiring regeneration) Wnt5a, Wnt5b, and Wnt7a are increased in expression. As the muscle completes repair Wnt7b and Wnt3a are increased as well. This patterning of Wnt signalling expression in muscle cell repair induces the differentiation of the progenitor cells, which reduces the number of available satellite cells. Wnt plays a crucial role in satellite cell regulation and skeletal muscle aging and also regeneration. Wnts are known to active the expression of Myf5 and MyoD by Wnt1 and Wnt7a. Wnt4, Wnt5, and Wnt6 function to increase the expression of both of the regulatory factors but at a more subtle level. Additionally, MyoD increases Wnt3a when myoblasts undergo differentiation. Whether MyoD is activated by Wnt via cis-regulation direct targeting or through indirect physiological pathways remains to be elucidated.[13]
Coactivators and repressors
IFRD1 is a positive cofactor of MyoD, as it cooperates with MyoD at inducing the transcriptional activity of MEF2C (by displacing HDAC4 from MEF2C); moreover IFRD1 also represses the transcriptional activity of NF-κB, which is known to inhibit MyoD mRNA accumulation.[14][15]
NFATc1 is a transcription factor that regulates composition of fiber type and the fast-to-slow twitch transition resulting from aerobic exercise requires the expression of NFATc1. MyoD expression is a key transcription factor in fast twitch fibers which is inhibited by NFATc1 in oxidative fiber types. NFATc1 works to inhibit MyoD via a physical interaction with the MyoD N-terminal activation domain resulting in inhibited recruitment of the necessary transcriptional coactivator p300. NFATc1 physically disrupts the interaction between MyoD and p300. This establishes the molecular mechanism by which fiber types transition in vivo through exercise with opposing roles for NFATc1 and MyoD. NFATc1 controls this balance by physical inhibition of MyoD in slow-twitch muscle fiber types.[16]

The histone deacetyltransferase p300 functions with MyoD in an interaction that is essential for the myotube generation from fibroblasts that is mediated by MyoD. Recruitment of p300 is the rate-limiting process in the conversion of fibroblasts to myotubes.[17] In addition to p300, MyoD is also known to recruit Set7, H3K4me1, H3K27ac, and RNAP II to the enhancer that is bound with and this allows for the activation of muscle gene that is condition-specific and established by MyoD recruitment. Endogenous p300 though, is necessary for MyoD functioning by acting as an essential coactivator. MyoD associatively binds to the enhancer region in conjunction with a placeholding "putative pioneer factor" which helps to establish and maintain a both of them in a specific and inactive conformation. Upon the removal or inactivation on the placeholder protein bound to the enhancer, the recruitment of the additional group of transcription factors that help to positively regulate enhancer activity is permitted and this results in the MyoD-transcription factor-enhancer complex to assume a transcriptionally active state.
Interactions
MyoD has been shown to interact with:
- C-jun,[18]
- CREB-binding protein,[19][20]
- CSRP3,[21]
- Cyclin-dependent kinase 4,[22][23]
- Cyclin-dependent kinase inhibitor 1C,[24]
- EP300,[20][25]
- HDAC1,[26][27]
- ID1,[28][29][30][31][32][33]
- ID2,[29]
- MDFI,[34]
- MOS,[35]
- Retinoblastoma protein,[27][36]
- Retinoid X receptor alpha[37]
- STAT3,[38] and
- TCF3.[29][39]
References
- ^ "Entrez Gene: MYOD1 myogenic differentiation 1".
- ^ Ehlers ML, Celona B, Black BL (Sep 2014). "NFATc1 controls skeletal muscle fiber type and is a negative regulator of MyoD activity". Cell Reports. 8 (6): 1639–1648. doi:10.1016/j.celrep.2014.08.035. PMID 25242327.
- ^ Tapscott SJ, Davis RL, Thayer MJ, Cheng PF, Weintraub H, Lassar AB (Oct 1988). "MyoD1: a nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts". Science. 242 (4877): 405–511. doi:10.1126/science.3175662. PMID 3175662.
- ^ Davis RL, Cheng PF, Lassar AB, Thayer M, Tapscott S, Weintraub H (1989). "MyoD and achaete-scute: 4-5 amino acids distinguishes myogenesis from neurogenesis". Princess Takamatsu Symposia. 20: 267–278. PMID 2562185.
- ^ Fong, A; Tapscott, S (October 2014). "Skeletal muscle programming and re-programming". Current Opinion in Genetics & Development. 23 (5): 568–573. doi:10.1016/j.gde.2013.05.002. PMID 23756045.
- ^ Singh K, Cassano M, Planet E, Sebastian S, Jang SM, Sohi G, Faralli H, Choi J, Youn HD, Dilworth FJ, Trono D (Mar 2015). "A KAP1 phosphorylation switch controls MyoD function during skeletal muscle differentiation". Genes & Development. 29 (5): 513–525. doi:10.1101/gad254532.114. PMID 25737281.
- ^ Buckingham, M; Rigby, P (February 2014). "Gene Regulatory Networks and Transcriptional Mechanisms that Control Myogenesis". Developmental Cell. 28 (3): 225–238. doi:10.1016/j.devcel.2013.12.020. PMID 24525185.
- ^ Song YJ, Choi JH, Lee H (Feb 2015). "Setdb1 Is Required for Myogenic Differentiation of C2C12 Myoblast Cells via Maintenance of MyoD Expression". Molecules and Cells. 38 (4). doi:10.14348/molcells.2015.2291. PMID 25715926.
- ^ Rajabi HN, Takahashi C, Ewen ME (Aug 2014). "Retinoblastoma protein and MyoD function together to effect the repression of Fra-1 and in turn cyclin D1 during terminal cell cycle arrest associated with myogenesis". The Journal of Biological Chemistry. 289 (34): 23417–23427. doi:10.1074/jbc.M113.532572. PMID 25006242.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Milewska, M; Grabiec, K; Grzelkowska-Kowalczyk, K (May 2014). "[Inteactions of proliferation and differentiation signaling pathways in myogenesis]". Postepy Hig Med Dosw. 8 (68): 516–526. doi:10.5604/17322693.1101617. PMID 24864103.
- ^ Pandur, P; Maurus, D; Kühl, M (October 2002). "Increasingly complex: new players enter the wnt signaling network". BioEssays. 24 (10): 881–884. doi:10.1002/bies.10164. PMID 12325120.
- ^ Pan YC, Wang XW, Teng HF, Wu YJ, Chang HC, Chen SL (Feb 2015). "Wnt3a signal pathways activate MyoD expression by targeting cis-elements inside and outside its distal enhancer". Bioscience Reports. 35: 1–12. doi:10.1042/BSR20140177. PMID 25651906.
- ^ Motohashi, N.; Asakura, Atsushi (January 2014). "Muscle satellite cell heterogeneity and self-renewal". Frontiers in Cell and Developmental Biology. 2 (1). doi:10.3389/fcell.2014.00001. PMID 25364710.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Micheli L, Leonardi L, Conti F, Buanne P, Canu N, Caruso M, Tirone F (March 2005). "PC4 coactivates MyoD by relieving the histone deacetylase 4-mediated inhibition of myocyte enhancer factor 2C". Mol. Cell. Biol. 25 (6): 2242–59. doi:10.1128/MCB.25.6.2242-2259.2005. PMC 1061592. PMID 15743821.
- ^ Micheli L, Leonardi L, Conti F, Maresca G, Colazingari S, Mattei E, Lira SA, Farioli-Vecchioli S, Caruso M, Tirone F (February 2011). "PC4/Tis7/IFRD1 stimulates skeletal muscle regeneration and is involved in myoblast differentiation as a regulator of MyoD and NF-kappaB". J. Biol. Chem. 286 (7): 5691–707. doi:10.1074/jbc.M110.162842. PMC 3037682. PMID 21127072.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Federation, A; Bradner, J; Meissner, A (March 2014). "The use of small molecules in somatic-cell reprogramming". Trends in Cellular Biology. 24 (3): 179–187. doi:10.1016/j.tcb.2013.09.011. PMID 24183602.
- ^ Sartorelli, V; Huang, J; Hamamori, Y; Kedes, L (February 1997). "Molecular mechanisms of myogenic coactivation by p300: direct interaction with the activation domain of MyoD and with the MADS box of MEF2C". Molecular Cell Biology. 17 (2): 1010–1026. doi:10.1128/mcb.17.2.1010. PMC 231826. PMID 9001254.
- ^ Bengal E, Ransone L, Scharfmann R, Dwarki VJ, Tapscott SJ, Weintraub H, Verma IM (February 1992). "Functional antagonism between c-Jun and MyoD proteins: a direct physical association". Cell. 68 (3): 507–19. doi:10.1016/0092-8674(92)90187-h. PMID 1310896.
- ^ Polesskaya A, Naguibneva I, Duquet A, Bengal E, Robin P, Harel-Bellan A (August 2001). "Interaction between acetylated MyoD and the bromodomain of CBP and/or p300". Mol. Cell. Biol. 21 (16): 5312–20. doi:10.1128/MCB.21.16.5312-5320.2001. PMC 87255. PMID 11463815.
- ^ a b Sartorelli V, Huang J, Hamamori Y, Kedes L (February 1997). "Molecular mechanisms of myogenic coactivation by p300: direct interaction with the activation domain of MyoD and with the MADS box of MEF2C". Mol. Cell. Biol. 17 (2): 1010–26. doi:10.1128/mcb.17.2.1010. PMC 231826. PMID 9001254.
- ^ Kong Y, Flick MJ, Kudla AJ, Konieczny SF (August 1997). "Muscle LIM protein promotes myogenesis by enhancing the activity of MyoD". Mol. Cell. Biol. 17 (8): 4750–60. doi:10.1128/mcb.17.8.4750. PMC 232327. PMID 9234731.
- ^ Zhang JM, Zhao X, Wei Q, Paterson BM (December 1999). "Direct inhibition of G(1) cdk kinase activity by MyoD promotes myoblast cell cycle withdrawal and terminal differentiation". EMBO J. 18 (24): 6983–93. doi:10.1093/emboj/18.24.6983. PMC 1171761. PMID 10601020.
- ^ Zhang JM, Wei Q, Zhao X, Paterson BM (February 1999). "Coupling of the cell cycle and myogenesis through the cyclin D1-dependent interaction of MyoD with cdk4". EMBO J. 18 (4): 926–33. doi:10.1093/emboj/18.4.926. PMC 1171185. PMID 10022835.
- ^ Reynaud EG, Leibovitch MP, Tintignac LA, Pelpel K, Guillier M, Leibovitch SA (June 2000). "Stabilization of MyoD by direct binding to p57(Kip2)". J. Biol. Chem. 275 (25): 18767–76. doi:10.1074/jbc.M907412199. PMID 10764802.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Lau P, Bailey P, Dowhan DH, Muscat GE (January 1999). "Exogenous expression of a dominant negative RORalpha1 vector in muscle cells impairs differentiation: RORalpha1 directly interacts with p300 and myoD". Nucleic Acids Res. 27 (2): 411–20. doi:10.1093/nar/27.2.411. PMC 148194. PMID 9862959.
- ^ Puri PL, Iezzi S, Stiegler P, Chen TT, Schiltz RL, Muscat GE, Giordano A, Kedes L, Wang JY, Sartorelli V (October 2001). "Class I histone deacetylases sequentially interact with MyoD and pRb during skeletal myogenesis". Mol. Cell. 8 (4): 885–97. doi:10.1016/s1097-2765(01)00373-2. PMID 11684023.
- ^ a b Mal A, Sturniolo M, Schiltz RL, Ghosh MK, Harter ML (April 2001). "A role for histone deacetylase HDAC1 in modulating the transcriptional activity of MyoD: inhibition of the myogenic program". EMBO J. 20 (7): 1739–53. doi:10.1093/emboj/20.7.1739. PMC 145490. PMID 11285237.
- ^ Garkavtsev I, Kozin SV, Chernova O, Xu L, Winkler F, Brown E, Barnett GH, Jain RK (March 2004). "The candidate tumour suppressor protein ING4 regulates brain tumour growth and angiogenesis". Nature. 428 (6980): 328–32. doi:10.1038/nature02329. PMID 15029197.
- ^ a b c Langlands K, Yin X, Anand G, Prochownik EV (August 1997). "Differential interactions of Id proteins with basic-helix-loop-helix transcription factors". J. Biol. Chem. 272 (32): 19785–93. doi:10.1074/jbc.272.32.19785. PMID 9242638.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Finkel T, Duc J, Fearon ER, Dang CV, Tomaselli GF (January 1993). "Detection and modulation in vivo of helix-loop-helix protein-protein interactions". J. Biol. Chem. 268 (1): 5–8. PMID 8380166.
- ^ Gupta K, Anand G, Yin X, Grove L, Prochownik EV (March 1998). "Mmip1: a novel leucine zipper protein that reverses the suppressive effects of Mad family members on c-myc". Oncogene. 16 (9): 1149–59. doi:10.1038/sj.onc.1201634. PMID 9528857.
- ^ McLoughlin P, Ehler E, Carlile G, Licht JD, Schäfer BW (October 2002). "The LIM-only protein DRAL/FHL2 interacts with and is a corepressor for the promyelocytic leukemia zinc finger protein". J. Biol. Chem. 277 (40): 37045–53. doi:10.1074/jbc.M203336200. PMID 12145280.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Ling MT, Chiu YT, Lee TK, Leung SC, Fung MK, Wang X, Wong KF, Wong YC (September 2008). "Id-1 induces proteasome-dependent degradation of the HBX protein". J. Mol. Biol. 382 (1): 34–43. doi:10.1016/j.jmb.2007.06.020. PMID 18674781.
- ^ Chen CM, Kraut N, Groudine M, Weintraub H (September 1996). "I-mf, a novel myogenic repressor, interacts with members of the MyoD family". Cell. 86 (5): 731–41. doi:10.1016/s0092-8674(00)80148-8. PMID 8797820.
- ^ Lenormand JL, Benayoun B, Guillier M, Vandromme M, Leibovitch MP, Leibovitch SA (February 1997). "Mos activates myogenic differentiation by promoting heterodimerization of MyoD and E12 proteins". Mol. Cell. Biol. 17 (2): 584–93. doi:10.1128/mcb.17.2.584. PMC 231783. PMID 9001211.
- ^ Gu W, Schneider JW, Condorelli G, Kaushal S, Mahdavi V, Nadal-Ginard B (February 1993). "Interaction of myogenic factors and the retinoblastoma protein mediates muscle cell commitment and differentiation". Cell. 72 (3): 309–24. doi:10.1016/0092-8674(93)90110-c. PMID 8381715.
- ^ Froeschlé A, Alric S, Kitzmann M, Carnac G, Auradé F, Rochette-Egly C, Bonnieu A (July 1998). "Retinoic acid receptors and muscle b-HLH proteins: partners in retinoid-induced myogenesis". Oncogene. 16 (26): 3369–78. doi:10.1038/sj.onc.1201894. PMID 9692544.
- ^ Kataoka Y, Matsumura I, Ezoe S, Nakata S, Takigawa E, Sato Y, Kawasaki A, Yokota T, Nakajima K, Felsani A, Kanakura Y (November 2003). "Reciprocal inhibition between MyoD and STAT3 in the regulation of growth and differentiation of myoblasts". J. Biol. Chem. 278 (45): 44178–87. doi:10.1074/jbc.M304884200. PMID 12947115.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Maleki SJ, Royer CA, Hurlburt BK (June 1997). "MyoD-E12 heterodimers and MyoD-MyoD homodimers are equally stable". Biochemistry. 36 (22): 6762–7. doi:10.1021/bi970262m. PMID 9184158.
External links
- MyoD+Protein at the U.S. National Library of Medicine Medical Subject Headings (MeSH)

